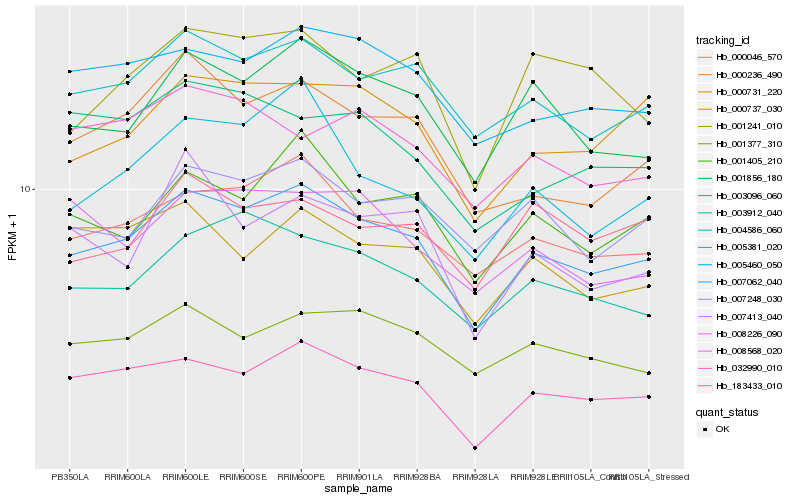
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007062_040 |
0.0 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 2 |
Hb_032990_010 |
0.045192145 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 3 |
Hb_000046_570 |
0.0497235565 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 4 |
Hb_004586_060 |
0.0521944959 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 5 |
Hb_001856_180 |
0.0583712938 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 6 |
Hb_001405_210 |
0.0604712223 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 7 |
Hb_005460_050 |
0.0611509094 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 8 |
Hb_000731_220 |
0.0617165241 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |
| 9 |
Hb_003912_040 |
0.0631917949 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 10 |
Hb_007248_030 |
0.0654428512 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 11 |
Hb_005381_020 |
0.0685000456 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 12 |
Hb_000236_490 |
0.0685277445 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 13 |
Hb_001377_310 |
0.0691355721 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 14 |
Hb_000737_030 |
0.0724149371 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001241_010 |
0.0767425383 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_008226_090 |
0.0772876459 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008568_020 |
0.0776729631 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 18 |
Hb_003096_060 |
0.0779655431 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 19 |
Hb_007413_040 |
0.0782054625 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 20 |
Hb_183433_010 |
0.0783328241 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |