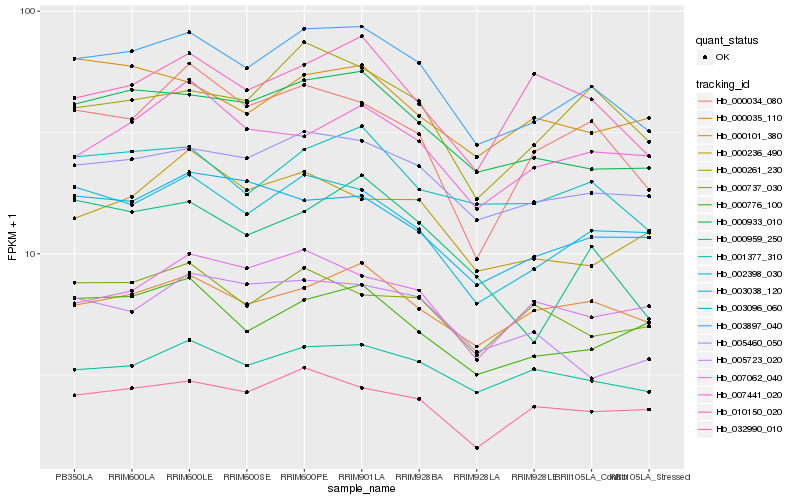
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003897_040 |
0.0 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 2 |
Hb_000933_010 |
0.055352259 |
- |
- |
- |
| 3 |
Hb_005460_050 |
0.0567693456 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 4 |
Hb_003038_120 |
0.0655448578 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 5 |
Hb_002398_030 |
0.0682995192 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000261_230 |
0.0752891313 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 7 |
Hb_032990_010 |
0.0767987237 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 8 |
Hb_001377_310 |
0.0788477466 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 9 |
Hb_000035_110 |
0.0792782868 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 10 |
Hb_000776_100 |
0.0818557253 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 11 |
Hb_000034_080 |
0.0825948234 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003096_060 |
0.0841825086 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 13 |
Hb_000737_030 |
0.0851747791 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000959_250 |
0.0862093807 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 15 |
Hb_007062_040 |
0.0896539493 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 16 |
Hb_007441_020 |
0.0902462893 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 17 |
Hb_000236_490 |
0.0909686203 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 18 |
Hb_010150_020 |
0.0914866363 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 19 |
Hb_000101_380 |
0.0919156592 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_005723_020 |
0.0923496703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |