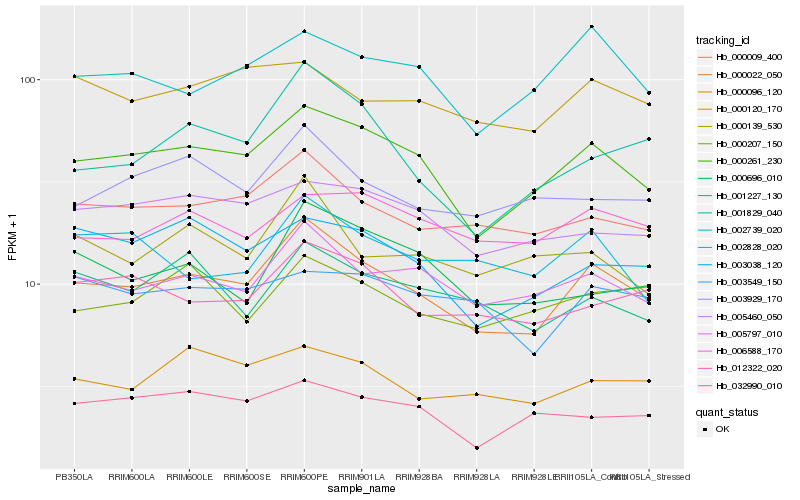
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_050 |
0.0 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 2 |
Hb_000261_230 |
0.0606325196 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 3 |
Hb_005797_010 |
0.0799041168 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000009_400 |
0.0844698494 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 5 |
Hb_012322_020 |
0.0950260983 |
- |
- |
- |
| 6 |
Hb_001829_040 |
0.0977743444 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 7 |
Hb_003929_170 |
0.0994832786 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 8 |
Hb_002739_020 |
0.1007989757 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 9 |
Hb_000096_120 |
0.1037034492 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 10 |
Hb_005460_050 |
0.1051504521 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 11 |
Hb_000139_530 |
0.1056098716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000696_010 |
0.1061497932 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 13 |
Hb_003038_120 |
0.1064340131 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 14 |
Hb_000207_150 |
0.1067935346 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 15 |
Hb_001227_130 |
0.1070779969 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 16 |
Hb_000120_170 |
0.1079353152 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 17 |
Hb_006588_170 |
0.1087197631 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 18 |
Hb_002828_020 |
0.1110616106 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_003549_150 |
0.1114273076 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 20 |
Hb_032990_010 |
0.1117418114 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |