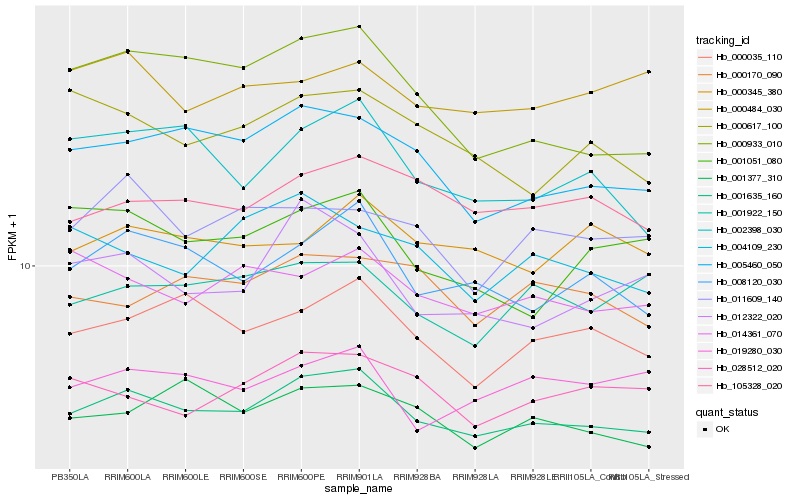
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001635_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 2 |
Hb_005460_050 |
0.0563335968 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 3 |
Hb_008120_030 |
0.0570080965 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 4 |
Hb_000035_110 |
0.0610885774 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 5 |
Hb_105328_020 |
0.0620158195 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 6 |
Hb_001377_310 |
0.0652577543 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 7 |
Hb_002398_030 |
0.0681680191 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_014361_070 |
0.0691535331 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 9 |
Hb_001922_150 |
0.0693121813 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_028512_020 |
0.0709183973 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000345_380 |
0.0712682041 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 12 |
Hb_000617_100 |
0.0714161737 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 13 |
Hb_000170_090 |
0.0716075144 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 14 |
Hb_011609_140 |
0.0718800692 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 15 |
Hb_019280_030 |
0.0725705837 |
- |
- |
- |
| 16 |
Hb_012322_020 |
0.073336992 |
- |
- |
- |
| 17 |
Hb_000484_030 |
0.0755151778 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 18 |
Hb_000933_010 |
0.0756350805 |
- |
- |
- |
| 19 |
Hb_001051_080 |
0.0757428828 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 20 |
Hb_004109_230 |
0.0759827961 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |