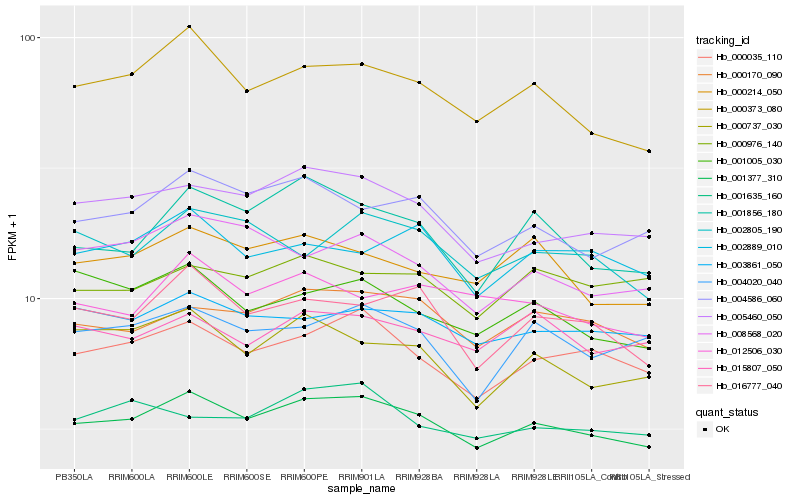
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_310 |
0.0 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 2 |
Hb_000035_110 |
0.0474938008 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 3 |
Hb_000170_090 |
0.0486646692 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 4 |
Hb_005460_050 |
0.0505463934 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 5 |
Hb_000373_080 |
0.0514778307 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 6 |
Hb_003861_050 |
0.0553062023 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 7 |
Hb_004586_060 |
0.0561162091 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 8 |
Hb_016777_040 |
0.0584572949 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 9 |
Hb_000214_050 |
0.060084502 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_015807_050 |
0.0616676046 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 11 |
Hb_004020_040 |
0.0631038915 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002889_010 |
0.0638738368 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 13 |
Hb_001635_160 |
0.0652577543 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 14 |
Hb_001005_030 |
0.0653397255 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_001856_180 |
0.0661849743 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 16 |
Hb_000737_030 |
0.0663102461 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008568_020 |
0.066620107 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 18 |
Hb_002805_190 |
0.0666971424 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 19 |
Hb_012506_030 |
0.0667610241 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 20 |
Hb_000976_140 |
0.0669565942 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |