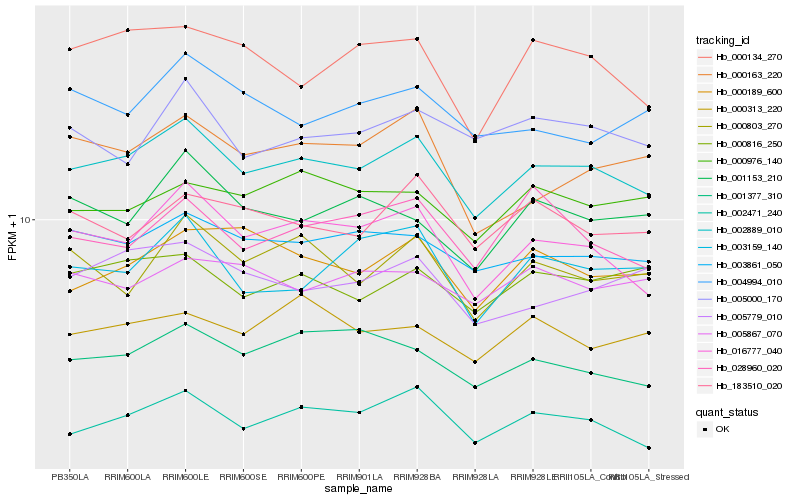
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002889_010 |
0.0 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 2 |
Hb_016777_040 |
0.0471222948 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 3 |
Hb_003861_050 |
0.0537409948 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 4 |
Hb_002471_240 |
0.0540659029 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001377_310 |
0.0638738368 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 6 |
Hb_000816_250 |
0.064036986 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000134_270 |
0.0662091905 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 8 |
Hb_005779_010 |
0.0674454223 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_000803_270 |
0.0675050364 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 10 |
Hb_183510_020 |
0.0676688342 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 11 |
Hb_000189_600 |
0.0683731704 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_003159_140 |
0.0686874918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000976_140 |
0.0695448607 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005000_170 |
0.06981731 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 15 |
Hb_000163_220 |
0.0708711893 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 16 |
Hb_028960_020 |
0.0715829279 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001153_210 |
0.0718505737 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000313_220 |
0.0720356887 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005867_070 |
0.0722866878 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_004994_010 |
0.0726950213 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |