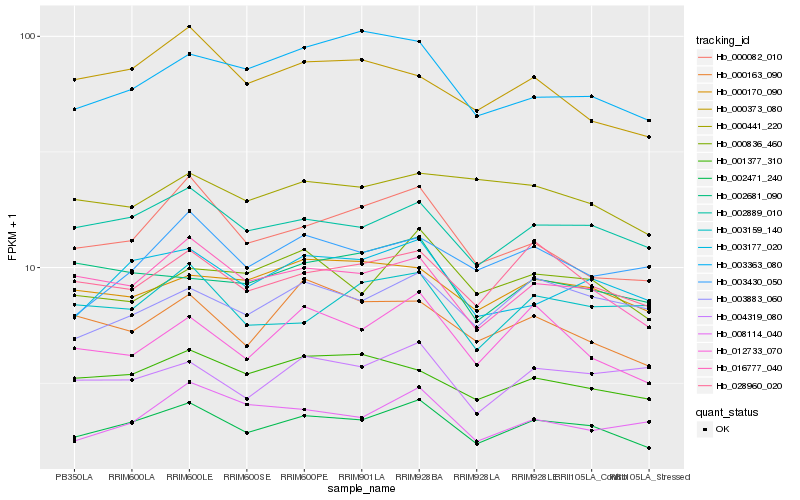
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_240 |
0.0 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002889_010 |
0.0540659029 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 3 |
Hb_016777_040 |
0.0579544146 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 4 |
Hb_003177_020 |
0.0653216855 |
- |
- |
diphthine synthase, putative [Ricinus communis] |
| 5 |
Hb_028960_020 |
0.0666742163 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003883_060 |
0.0741686136 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001377_310 |
0.0757441419 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 8 |
Hb_000082_010 |
0.0768282843 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 9 |
Hb_012733_070 |
0.077067482 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000170_090 |
0.0774485151 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 11 |
Hb_000836_460 |
0.0792743645 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 12 |
Hb_000163_090 |
0.0838639926 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 13 |
Hb_003430_050 |
0.0842727822 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 14 |
Hb_003363_080 |
0.0845428676 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 15 |
Hb_008114_040 |
0.0854521589 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 16 |
Hb_004319_080 |
0.0854723198 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 17 |
Hb_003159_140 |
0.0856070702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002681_090 |
0.0856290834 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000441_220 |
0.0860311818 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000373_080 |
0.0862466231 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |