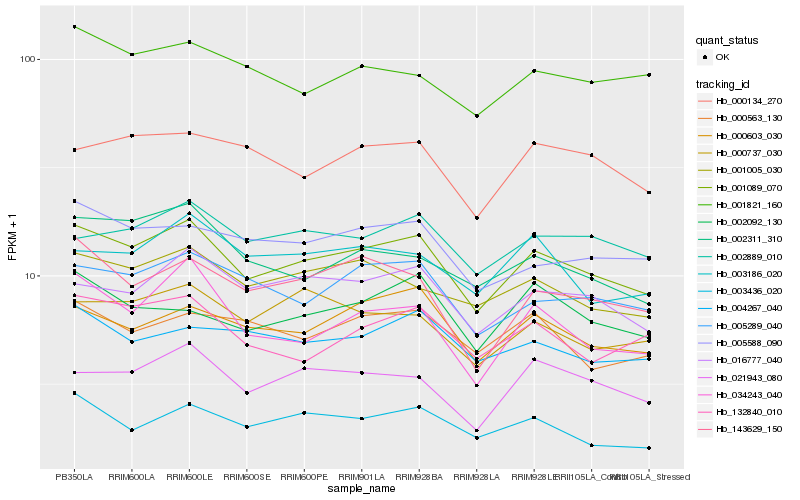
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_070 |
0.0 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 2 |
Hb_016777_040 |
0.0667899347 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 3 |
Hb_005289_040 |
0.0671025251 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 4 |
Hb_000603_030 |
0.0682416361 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 5 |
Hb_001005_030 |
0.0696653025 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_021943_080 |
0.0713491954 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005588_090 |
0.0736202423 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 8 |
Hb_002889_010 |
0.0745206131 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 9 |
Hb_143629_150 |
0.0754141782 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 10 |
Hb_003436_020 |
0.075624968 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 11 |
Hb_132840_010 |
0.0767859308 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_034243_040 |
0.076935919 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 13 |
Hb_002311_310 |
0.0780928955 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 14 |
Hb_000737_030 |
0.0786458955 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000134_270 |
0.078683283 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 16 |
Hb_004267_040 |
0.0793951979 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 17 |
Hb_002092_130 |
0.0795873438 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 18 |
Hb_003186_020 |
0.0803273413 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000563_130 |
0.0816068636 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 20 |
Hb_001821_160 |
0.0821578456 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |