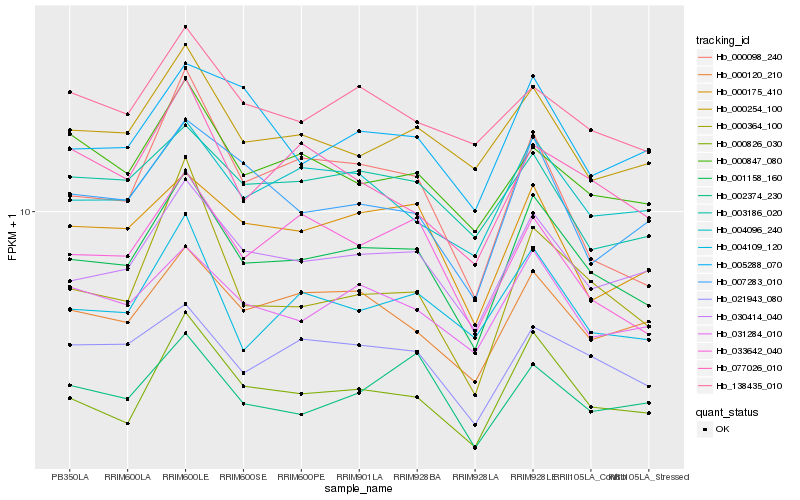
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_160 |
0.0 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_030414_040 |
0.0568874629 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000826_030 |
0.0614575772 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 4 |
Hb_021943_080 |
0.0698076904 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_031284_010 |
0.0708427659 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 6 |
Hb_000120_210 |
0.0719753337 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 7 |
Hb_004096_240 |
0.0732402743 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 8 |
Hb_004109_120 |
0.076685872 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000175_410 |
0.0770958424 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000254_100 |
0.0789461218 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 11 |
Hb_007283_010 |
0.0791689107 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 12 |
Hb_033642_040 |
0.0818735715 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002374_230 |
0.0843144335 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 14 |
Hb_000847_080 |
0.0845522526 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_003186_020 |
0.0847897155 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000098_240 |
0.0861413676 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 17 |
Hb_000364_100 |
0.0865377992 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 18 |
Hb_077026_010 |
0.0871288187 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 19 |
Hb_138435_010 |
0.0876816554 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 20 |
Hb_005288_070 |
0.0876920765 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |