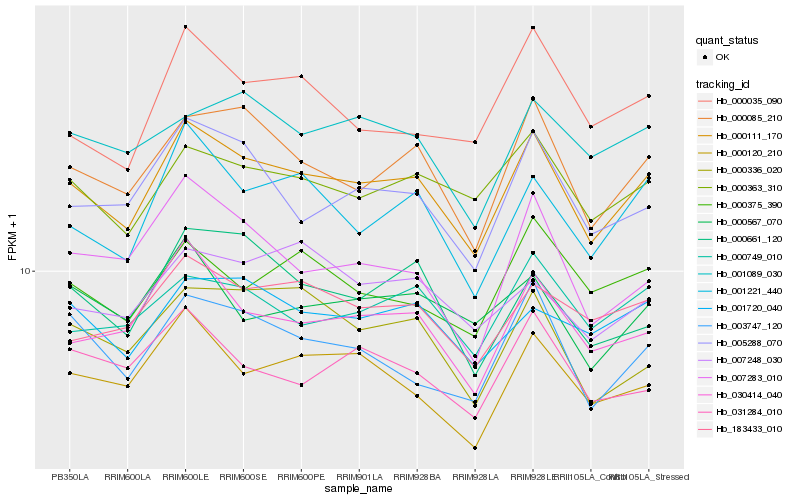
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000085_210 |
0.0554585708 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 3 |
Hb_000363_310 |
0.0627577296 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 4 |
Hb_001221_440 |
0.06389163 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_030414_040 |
0.064162499 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005288_070 |
0.0661241281 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_183433_010 |
0.0679350094 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 8 |
Hb_000120_210 |
0.068540979 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 9 |
Hb_003747_120 |
0.0692572414 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000749_010 |
0.0697197596 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001720_040 |
0.0702189562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001089_030 |
0.0754342541 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 13 |
Hb_000375_390 |
0.0756323437 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 14 |
Hb_000336_020 |
0.0757225285 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 15 |
Hb_000567_070 |
0.0760395967 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 16 |
Hb_000661_120 |
0.0768734729 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 17 |
Hb_007283_010 |
0.0772577972 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 18 |
Hb_000035_090 |
0.077279102 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 19 |
Hb_031284_010 |
0.0780977384 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 20 |
Hb_007248_030 |
0.0784427348 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |