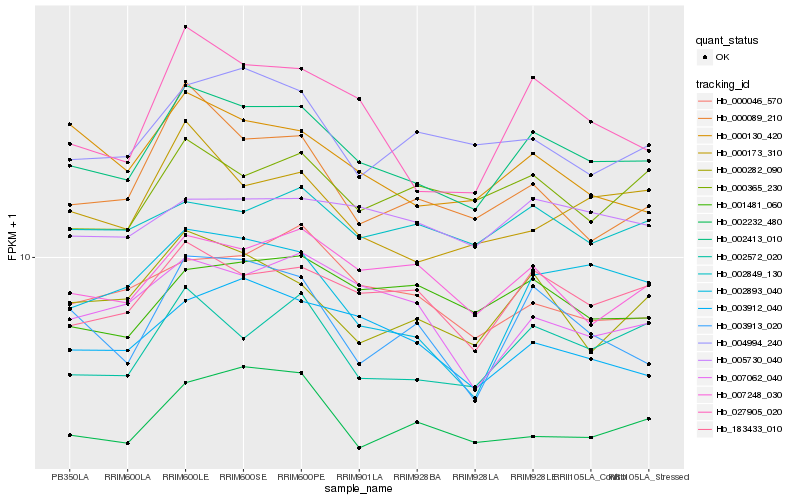
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002413_010 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_183433_010 |
0.0580886186 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 3 |
Hb_027905_020 |
0.0661414913 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 4 |
Hb_002572_020 |
0.0677503808 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 5 |
Hb_007248_030 |
0.0706523234 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 6 |
Hb_000282_090 |
0.0744801923 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_003912_040 |
0.0747582746 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 8 |
Hb_000173_310 |
0.0759425005 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000130_420 |
0.076498495 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 10 |
Hb_001481_060 |
0.0788552914 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002893_040 |
0.0789499233 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004994_240 |
0.0791788393 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 13 |
Hb_002232_480 |
0.079334521 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 14 |
Hb_000046_570 |
0.0794135996 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 15 |
Hb_000089_210 |
0.0799008808 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_007062_040 |
0.0800780719 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 17 |
Hb_000365_230 |
0.0806786906 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 18 |
Hb_005730_040 |
0.0807070417 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 19 |
Hb_002849_130 |
0.0807533352 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 20 |
Hb_003913_020 |
0.0807759428 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |