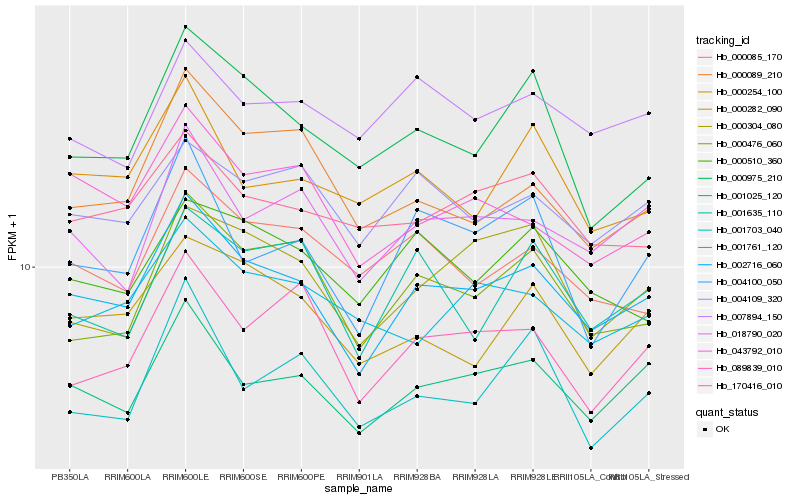
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002716_060 |
0.0 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 2 |
Hb_001025_120 |
0.0516376037 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_018790_020 |
0.0678994156 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000476_060 |
0.072877974 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_004109_320 |
0.0730652486 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 6 |
Hb_043792_010 |
0.0733778287 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000282_090 |
0.0739423292 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 8 |
Hb_170416_010 |
0.0757669425 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 9 |
Hb_004100_050 |
0.0777364858 |
- |
- |
PREDICTED: mitochondrial-processing peptidase subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_089839_010 |
0.0795454817 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 11 |
Hb_000510_360 |
0.0797637202 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 12 |
Hb_001761_120 |
0.0797805044 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 13 |
Hb_000975_210 |
0.0803948648 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 14 |
Hb_007894_150 |
0.0805734102 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 15 |
Hb_000089_210 |
0.0810597298 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_000085_170 |
0.0817221437 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000304_080 |
0.082117177 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000254_100 |
0.0821511877 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 19 |
Hb_001635_110 |
0.0842973626 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 20 |
Hb_001703_040 |
0.0863923397 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |