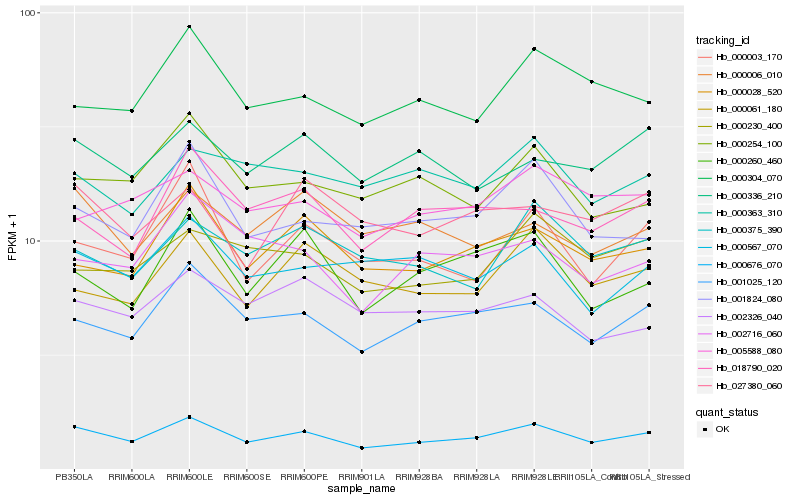
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 2 |
Hb_001025_120 |
0.069163988 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_000028_520 |
0.085955944 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 4 |
Hb_000336_210 |
0.0904887222 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001824_080 |
0.0911927109 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 6 |
Hb_005588_080 |
0.0914225348 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 7 |
Hb_000567_070 |
0.0923738748 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 8 |
Hb_000375_390 |
0.0924761148 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 9 |
Hb_000304_070 |
0.0927083253 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 10 |
Hb_000260_460 |
0.0928224888 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000006_010 |
0.0930828451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_018790_020 |
0.0930992933 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000061_180 |
0.0933379369 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 14 |
Hb_000230_400 |
0.0937267077 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000254_100 |
0.0937533645 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 16 |
Hb_000003_170 |
0.0944744025 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 17 |
Hb_002716_060 |
0.0946736574 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 18 |
Hb_027380_060 |
0.0946994541 |
- |
- |
PREDICTED: putative ALA-interacting subunit 2 [Jatropha curcas] |
| 19 |
Hb_000363_310 |
0.096238705 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 20 |
Hb_002326_040 |
0.0969928471 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |