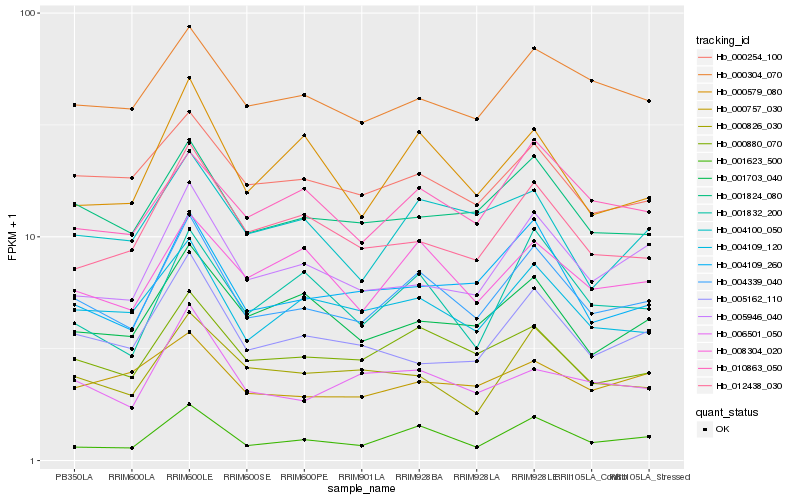
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004339_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 2 |
Hb_000880_070 |
0.0740006641 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 3 |
Hb_001824_080 |
0.0778625589 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 4 |
Hb_004109_120 |
0.0797334128 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004109_260 |
0.0830922929 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 6 |
Hb_001623_500 |
0.0861044806 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 7 |
Hb_005946_040 |
0.0893841654 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000304_070 |
0.0895501112 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 9 |
Hb_008304_020 |
0.0909982488 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 10 |
Hb_000254_100 |
0.0910891107 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 11 |
Hb_000579_080 |
0.0945596451 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 12 |
Hb_010863_050 |
0.0946584693 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 13 |
Hb_001703_040 |
0.0951593623 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 14 |
Hb_012438_030 |
0.0958438178 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 15 |
Hb_004100_050 |
0.0977020877 |
- |
- |
PREDICTED: mitochondrial-processing peptidase subunit alpha-like [Jatropha curcas] |
| 16 |
Hb_005162_110 |
0.0981498492 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 17 |
Hb_001832_200 |
0.0988594824 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006501_050 |
0.0991987184 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 19 |
Hb_000826_030 |
0.0992343951 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 20 |
Hb_000757_030 |
0.1028042494 |
- |
- |
radical sam protein, putative [Ricinus communis] |