| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
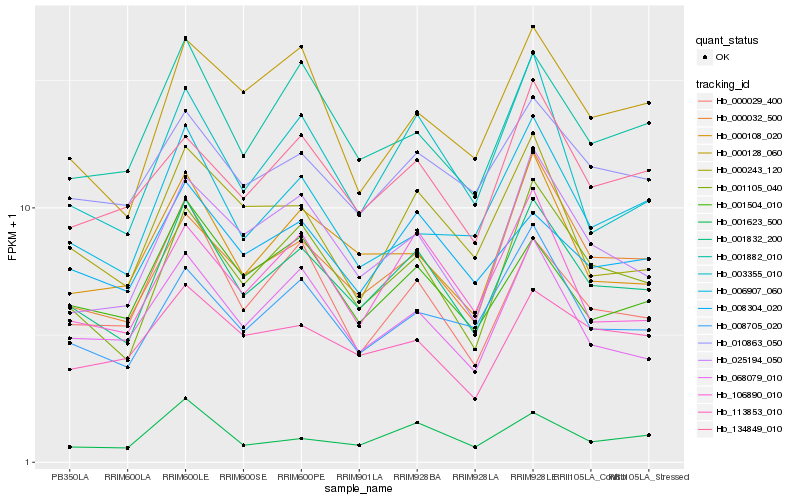
Hb_001832_200 |
0.0 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001105_040 |
0.0615372227 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_001882_010 |
0.0810015027 |
- |
- |
- |
| 4 |
Hb_025194_050 |
0.0811768449 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 5 |
Hb_010863_050 |
0.0849021106 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 6 |
Hb_000108_020 |
0.0859207163 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 7 |
Hb_068079_010 |
0.0890238448 |
- |
- |
- |
| 8 |
Hb_008705_020 |
0.0890532353 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_006907_060 |
0.0897328229 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_001504_010 |
0.0912325018 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 11 |
Hb_000032_500 |
0.0913817454 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 12 |
Hb_000029_400 |
0.091668041 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008304_020 |
0.0918489025 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 14 |
Hb_003355_010 |
0.0921770036 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 15 |
Hb_106890_010 |
0.093505668 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_134849_010 |
0.0948572371 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_001623_500 |
0.0949756938 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 18 |
Hb_000243_120 |
0.0972259377 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000128_060 |
0.0974335508 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 20 |
Hb_113853_010 |
0.0980064567 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |