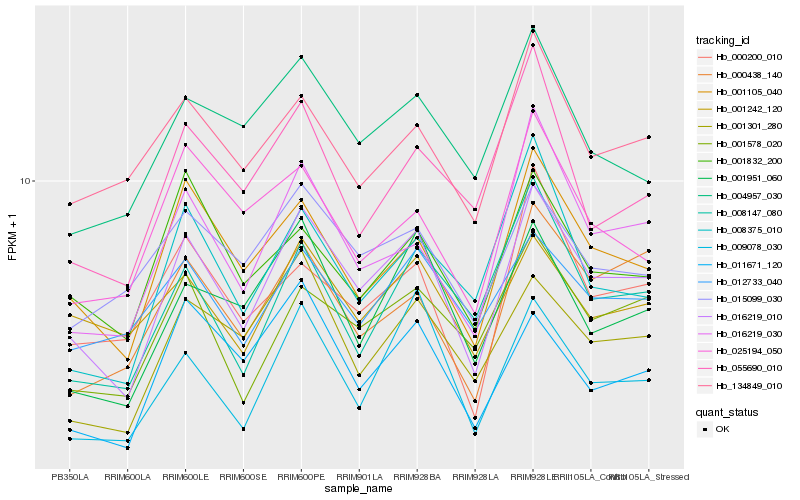
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016219_010 |
0.0 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 2 |
Hb_001105_040 |
0.0911159776 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_016219_030 |
0.0996428787 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_001578_020 |
0.1081425756 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 5 |
Hb_001951_060 |
0.109047445 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 6 |
Hb_001301_280 |
0.1107472535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001242_120 |
0.1162742273 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 8 |
Hb_000438_140 |
0.1165501316 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 9 |
Hb_012733_040 |
0.1177176981 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 10 |
Hb_001832_200 |
0.1178303594 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008375_010 |
0.1178767672 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 12 |
Hb_008147_080 |
0.1195116756 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 13 |
Hb_015099_030 |
0.1203643553 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 14 |
Hb_000200_010 |
0.1209477108 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 15 |
Hb_004957_030 |
0.1213373874 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 16 |
Hb_134849_010 |
0.1239862298 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_055690_010 |
0.124344707 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_011671_120 |
0.1250269192 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 19 |
Hb_025194_050 |
0.1251680025 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009078_030 |
0.1254601114 |
- |
- |
hypothetical protein PRUPE_ppa016992mg, partial [Prunus persica] |