| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
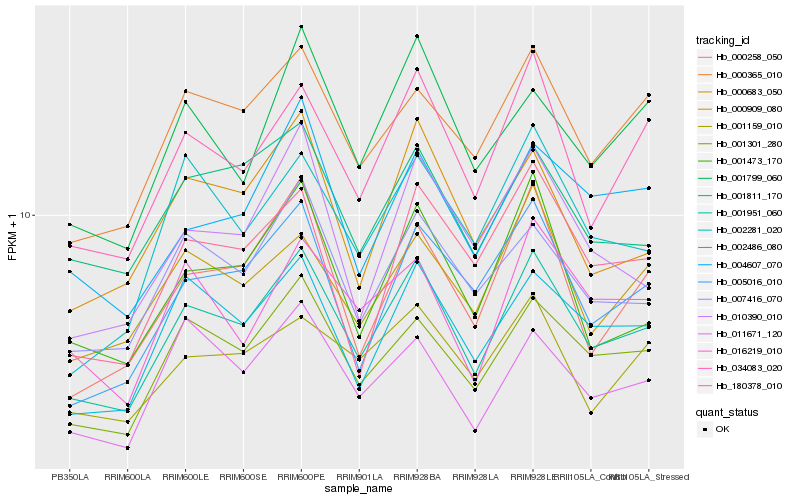
Hb_001951_060 |
0.0 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 2 |
Hb_001301_280 |
0.0737087407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_034083_020 |
0.0819910457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002486_080 |
0.0838307405 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 5 |
Hb_007416_070 |
0.0912252639 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 6 |
Hb_000365_010 |
0.0934756101 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 7 |
Hb_005016_010 |
0.0958897709 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 8 |
Hb_000683_050 |
0.0981341709 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 9 |
Hb_001473_170 |
0.0991906976 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 10 |
Hb_000258_050 |
0.1009148332 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002281_020 |
0.1019786508 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 12 |
Hb_010390_010 |
0.1023669871 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 13 |
Hb_000909_080 |
0.1038830451 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 14 |
Hb_180378_010 |
0.1049329367 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 15 |
Hb_001811_170 |
0.1050583593 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_011671_120 |
0.105867152 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 17 |
Hb_001799_060 |
0.1060573548 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 18 |
Hb_001159_010 |
0.1066993107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004607_070 |
0.1067606016 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 20 |
Hb_016219_010 |
0.109047445 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |