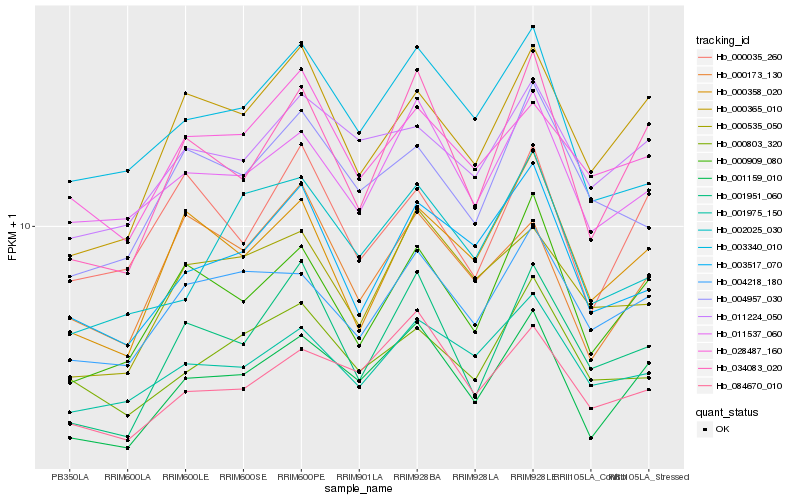
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001159_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_034083_020 |
0.0714973989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_084670_010 |
0.0860087409 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000909_080 |
0.0917889619 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 5 |
Hb_000365_010 |
0.0999766093 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 6 |
Hb_004218_180 |
0.1009461507 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 7 |
Hb_011224_050 |
0.1010473118 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 8 |
Hb_011537_060 |
0.1015350067 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 9 |
Hb_001951_060 |
0.1066993107 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 10 |
Hb_001975_150 |
0.11087527 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 11 |
Hb_003517_070 |
0.1149441409 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 12 |
Hb_003340_010 |
0.1152008951 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 13 |
Hb_000358_020 |
0.1153369976 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 14 |
Hb_000535_050 |
0.1154336284 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 15 |
Hb_002025_030 |
0.1161298566 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 16 |
Hb_000035_260 |
0.1210525988 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 17 |
Hb_000173_130 |
0.1215513751 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000803_320 |
0.12197401 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 19 |
Hb_028487_160 |
0.1241813443 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 20 |
Hb_004957_030 |
0.12677202 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |