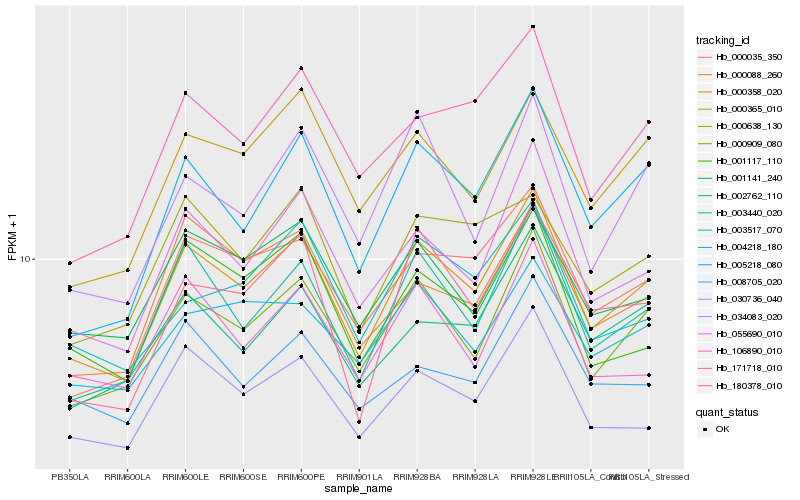
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000358_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 2 |
Hb_000909_080 |
0.0575088062 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 3 |
Hb_055690_010 |
0.063096957 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003440_020 |
0.0672546522 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_005218_080 |
0.0689883538 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000035_350 |
0.0717616068 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_030736_040 |
0.0746127489 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 8 |
Hb_171718_010 |
0.0811151005 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 9 |
Hb_000365_010 |
0.0843995717 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 10 |
Hb_002762_110 |
0.0849254212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001141_240 |
0.0873592173 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001117_110 |
0.0893542278 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 13 |
Hb_000088_260 |
0.0895748466 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 14 |
Hb_106890_010 |
0.0900073853 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_004218_180 |
0.0902983252 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 16 |
Hb_000638_130 |
0.0919295285 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 17 |
Hb_034083_020 |
0.0926384149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_180378_010 |
0.0927619484 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 19 |
Hb_008705_020 |
0.0943021367 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_003517_070 |
0.0955081769 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |