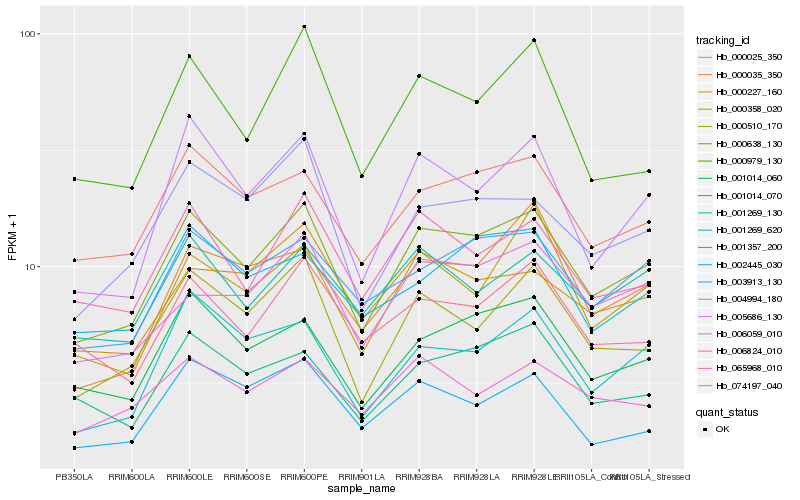
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000638_130 |
0.0 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 2 |
Hb_000025_350 |
0.0712790044 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001014_060 |
0.0745207058 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 4 |
Hb_005686_130 |
0.0753819113 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 5 |
Hb_000510_170 |
0.0755243119 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 6 |
Hb_003913_130 |
0.0787040828 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 7 |
Hb_065968_010 |
0.0821280813 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001269_620 |
0.0857827849 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 9 |
Hb_004994_180 |
0.0867505249 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 10 |
Hb_006059_010 |
0.0869978711 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 11 |
Hb_001269_130 |
0.0876626012 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 12 |
Hb_001014_070 |
0.0877767069 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 13 |
Hb_074197_040 |
0.0904321158 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 14 |
Hb_000358_020 |
0.0919295285 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 15 |
Hb_000979_130 |
0.0935556137 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_000035_350 |
0.0939008017 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001357_200 |
0.0943424142 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_002445_030 |
0.0946021183 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 19 |
Hb_000227_160 |
0.0947835197 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 20 |
Hb_006824_010 |
0.0950497059 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |