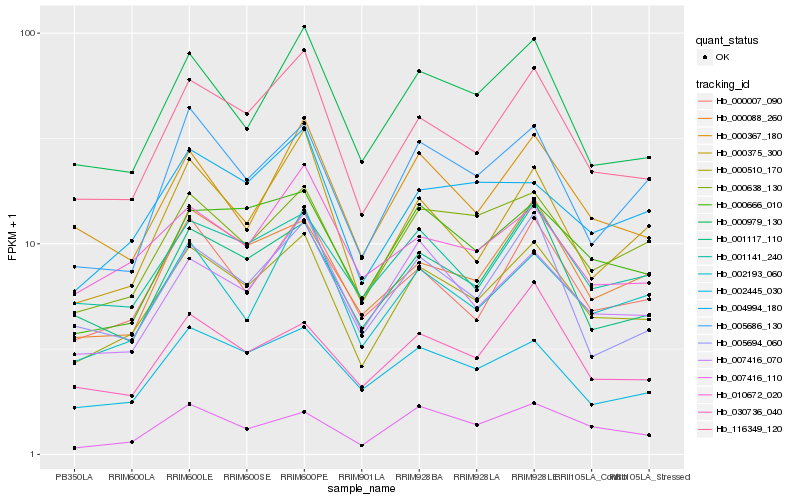
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_170 |
0.0 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 2 |
Hb_116349_120 |
0.058888892 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000638_130 |
0.0755243119 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 4 |
Hb_001141_240 |
0.0824309954 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000979_130 |
0.0849505549 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 6 |
Hb_000666_010 |
0.085790361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002445_030 |
0.0859300167 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 8 |
Hb_000088_260 |
0.0866401621 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 9 |
Hb_005686_130 |
0.0884478001 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 10 |
Hb_000007_090 |
0.0886911478 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 11 |
Hb_007416_070 |
0.0888672844 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 12 |
Hb_001117_110 |
0.0893622395 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 13 |
Hb_004994_180 |
0.091327214 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_000375_300 |
0.0929439546 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 15 |
Hb_000367_180 |
0.0954622961 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 16 |
Hb_002193_060 |
0.0955729176 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 17 |
Hb_007416_110 |
0.0958760026 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005694_060 |
0.0975793046 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 19 |
Hb_030736_040 |
0.0994919461 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010672_020 |
0.1006609169 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |