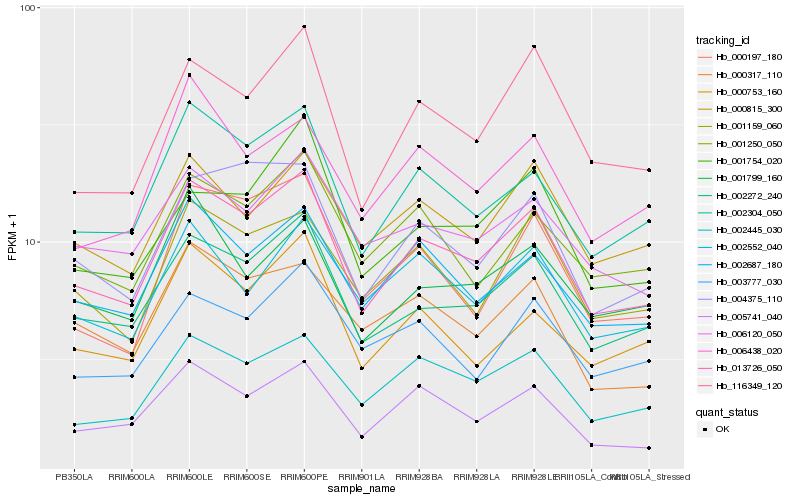
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013726_050 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 2 |
Hb_002272_240 |
0.0528730629 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_002304_050 |
0.0610535548 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 4 |
Hb_005741_040 |
0.0705863805 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 5 |
Hb_002687_180 |
0.0731071897 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 6 |
Hb_002445_030 |
0.0734457679 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 7 |
Hb_116349_120 |
0.0754334169 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_004375_110 |
0.0778621324 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 9 |
Hb_006120_050 |
0.0838735724 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 10 |
Hb_001250_050 |
0.0842019326 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001159_060 |
0.0842275549 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 12 |
Hb_000815_300 |
0.0867146881 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 13 |
Hb_000753_160 |
0.0878593315 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 14 |
Hb_000317_110 |
0.0902609721 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_002552_040 |
0.0904331451 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 16 |
Hb_006438_020 |
0.0923765768 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 17 |
Hb_001754_020 |
0.0925000296 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000197_180 |
0.0927967491 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 19 |
Hb_003777_030 |
0.093091065 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 20 |
Hb_001799_160 |
0.0943644257 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |