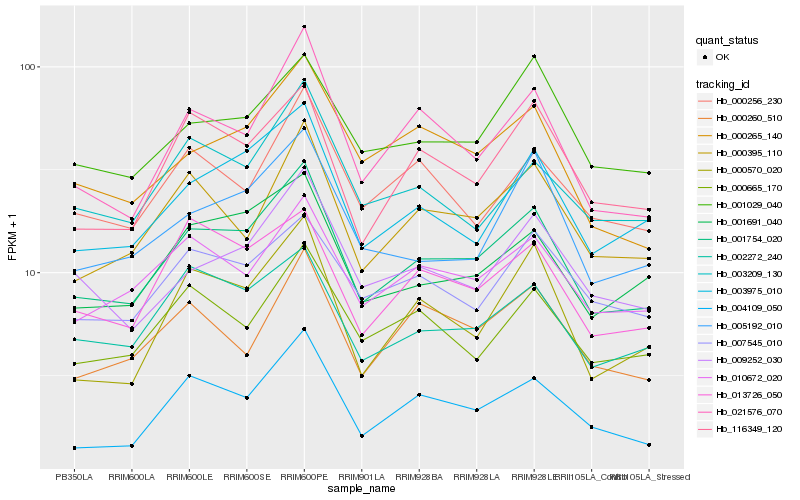
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_020 |
0.0 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 2 |
Hb_003975_010 |
0.0803126251 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 3 |
Hb_021576_070 |
0.0816075845 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 4 |
Hb_001691_040 |
0.0818545272 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 5 |
Hb_010672_020 |
0.0823459255 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_002272_240 |
0.0833327568 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_116349_120 |
0.0865949716 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_000395_110 |
0.086868074 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 9 |
Hb_000265_140 |
0.0891172616 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 10 |
Hb_009252_030 |
0.0897475406 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 11 |
Hb_004109_050 |
0.0921609972 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 12 |
Hb_005192_010 |
0.0922821884 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_013726_050 |
0.0925000296 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 14 |
Hb_001029_040 |
0.0937148232 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 15 |
Hb_000665_170 |
0.0953085574 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 16 |
Hb_007545_010 |
0.0966596976 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 17 |
Hb_003209_130 |
0.0967166363 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 18 |
Hb_000570_020 |
0.0974593807 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 19 |
Hb_000256_230 |
0.1005137216 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 20 |
Hb_000260_510 |
0.1023305461 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |