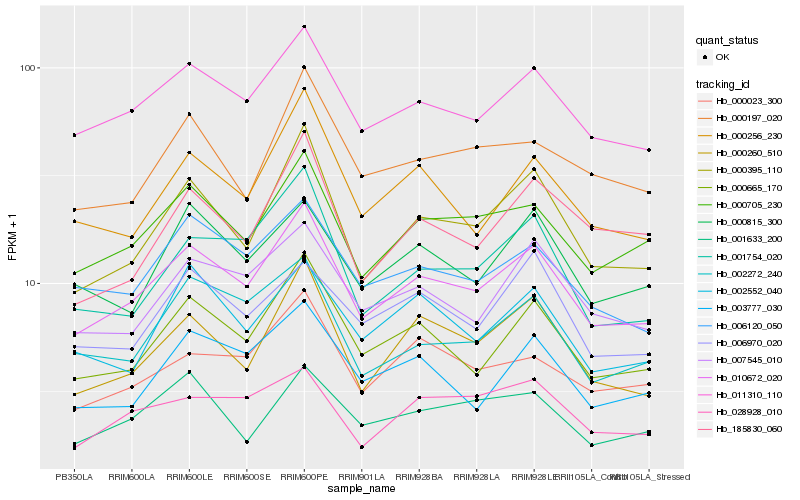
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010672_020 |
0.0 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_011310_110 |
0.0463836768 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000705_230 |
0.0594570516 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 4 |
Hb_000665_170 |
0.060362856 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 5 |
Hb_000395_110 |
0.0629576583 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 6 |
Hb_007545_010 |
0.0737106936 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 7 |
Hb_006120_050 |
0.0745467602 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 8 |
Hb_000260_510 |
0.0753861008 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000256_230 |
0.0783976072 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 10 |
Hb_000023_300 |
0.081428006 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 11 |
Hb_003777_030 |
0.0821980799 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 12 |
Hb_001754_020 |
0.0823459255 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 13 |
Hb_002272_240 |
0.083663956 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000197_020 |
0.0859647596 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 15 |
Hb_000815_300 |
0.0863918714 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 16 |
Hb_002552_040 |
0.0893349092 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 17 |
Hb_185830_060 |
0.0909842211 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 18 |
Hb_028928_010 |
0.0912307868 |
- |
- |
hypothetical protein CISIN_1g0030132mg, partial [Citrus sinensis] |
| 19 |
Hb_001633_200 |
0.0917405124 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 20 |
Hb_006970_020 |
0.091896063 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |