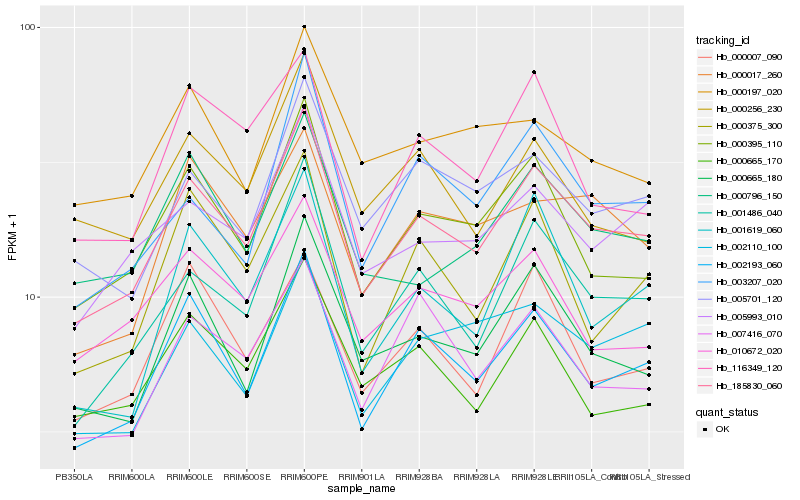
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185830_060 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 2 |
Hb_002193_060 |
0.0604174068 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 3 |
Hb_001486_040 |
0.0716217133 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000395_110 |
0.0766283918 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 5 |
Hb_005993_010 |
0.0802021786 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 6 |
Hb_000665_180 |
0.0823346623 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_005701_120 |
0.0830049005 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 8 |
Hb_000796_150 |
0.0891784095 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 9 |
Hb_010672_020 |
0.0909842211 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000017_260 |
0.0924545486 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 11 |
Hb_000665_170 |
0.0927164441 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 12 |
Hb_003207_020 |
0.095725762 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 13 |
Hb_007416_070 |
0.0958165008 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 14 |
Hb_002110_100 |
0.0959601882 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000007_090 |
0.0965624749 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 16 |
Hb_000197_020 |
0.0993098695 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 17 |
Hb_001619_060 |
0.0993197922 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 18 |
Hb_116349_120 |
0.0993339484 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000375_300 |
0.0996942277 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 20 |
Hb_000256_230 |
0.0998356732 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |