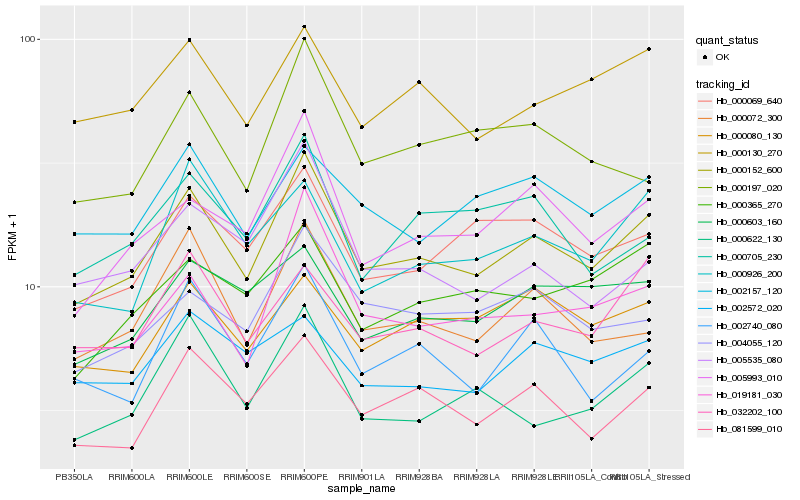
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_600 |
0.0 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 2 |
Hb_081599_010 |
0.0781600857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005993_010 |
0.0860880105 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 4 |
Hb_000072_300 |
0.0865047172 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 5 |
Hb_019181_030 |
0.0886556409 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 6 |
Hb_000130_270 |
0.0894949592 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000069_640 |
0.0900870906 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 8 |
Hb_032202_100 |
0.0914827067 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 9 |
Hb_002740_080 |
0.0927832674 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 10 |
Hb_002157_120 |
0.0945288909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004055_120 |
0.0948337634 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 12 |
Hb_000926_200 |
0.0972140333 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_000622_130 |
0.0987719431 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 14 |
Hb_005535_080 |
0.0988407537 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 15 |
Hb_000365_270 |
0.0991597324 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 16 |
Hb_000603_160 |
0.0991843943 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 17 |
Hb_000705_230 |
0.0995554701 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 18 |
Hb_002572_020 |
0.1020823377 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 19 |
Hb_000197_020 |
0.1027226166 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 20 |
Hb_000080_130 |
0.1027247026 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |