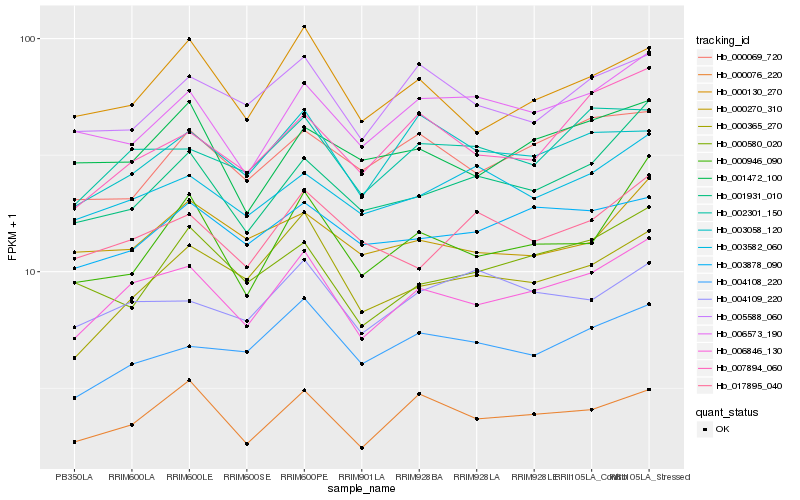
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_130 |
0.0 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 1-like [Jatropha curcas] |
| 2 |
Hb_002301_150 |
0.0686676768 |
- |
- |
Drought-responsive family protein [Theobroma cacao] |
| 3 |
Hb_000076_220 |
0.0751843411 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000130_270 |
0.0788494511 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_006573_190 |
0.0834767255 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 6 |
Hb_003582_060 |
0.0838433967 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001931_010 |
0.0842620442 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 8 |
Hb_004108_220 |
0.084769553 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003878_090 |
0.0857672584 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 10 |
Hb_007894_060 |
0.0859617435 |
- |
- |
PREDICTED: ubiquitin thioesterase OTU1 [Jatropha curcas] |
| 11 |
Hb_000365_270 |
0.0865913477 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 12 |
Hb_005588_060 |
0.0872946586 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 13 |
Hb_003058_120 |
0.0874123573 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 14 |
Hb_001472_100 |
0.0888828281 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 15 |
Hb_000946_090 |
0.0903820399 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_017895_040 |
0.0914965424 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 17 |
Hb_004109_220 |
0.0915693808 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 9-like [Populus euphratica] |
| 18 |
Hb_000270_310 |
0.091866786 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 19 |
Hb_000069_720 |
0.092013923 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 20 |
Hb_000580_020 |
0.0931164333 |
- |
- |
unnamed protein product [Vitis vinifera] |