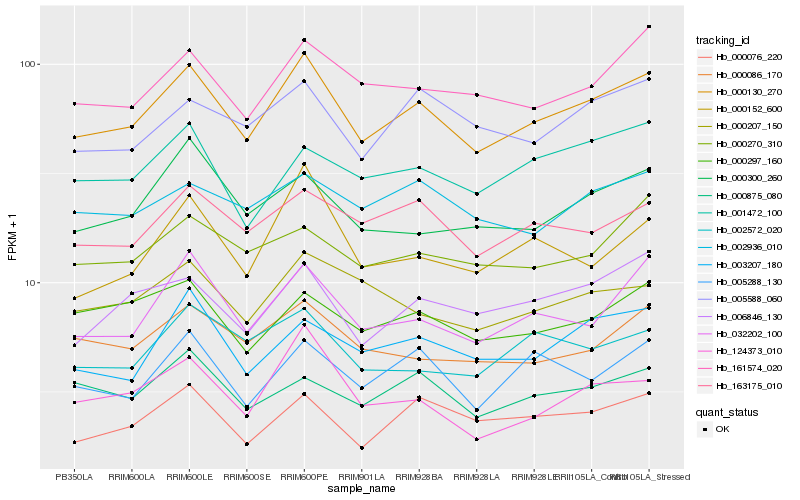
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_270 |
0.0 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_032202_100 |
0.0722914061 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_161574_020 |
0.0771823376 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 4 |
Hb_163175_010 |
0.0774232369 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 5 |
Hb_006846_130 |
0.0788494511 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 1-like [Jatropha curcas] |
| 6 |
Hb_000076_220 |
0.0800997353 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000086_170 |
0.0822742533 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 8 |
Hb_003207_180 |
0.0829293711 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005288_130 |
0.0835688146 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_005588_060 |
0.0837626995 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000300_260 |
0.084268184 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 12 |
Hb_000297_160 |
0.0844276925 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 13 |
Hb_001472_100 |
0.0862559009 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 14 |
Hb_000207_150 |
0.0865640932 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 15 |
Hb_000875_080 |
0.0870119238 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_000270_310 |
0.0877075669 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 17 |
Hb_002572_020 |
0.0884490451 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 18 |
Hb_002936_010 |
0.0887846785 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 19 |
Hb_000152_600 |
0.0894949592 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 20 |
Hb_124373_010 |
0.089502312 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |