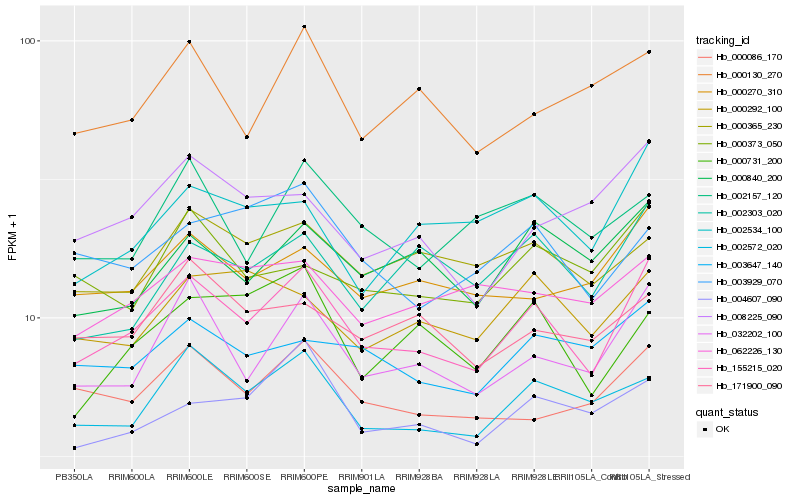
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155215_020 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Populus euphratica] |
| 2 |
Hb_032202_100 |
0.075398351 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_002572_020 |
0.0831486445 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 4 |
Hb_000086_170 |
0.0912144851 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 5 |
Hb_000270_310 |
0.092450234 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 6 |
Hb_004607_090 |
0.094256948 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 7 |
Hb_000840_200 |
0.0959333225 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_003647_140 |
0.0986327621 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 9 |
Hb_000365_230 |
0.0989851044 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 10 |
Hb_002534_100 |
0.099866376 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000373_050 |
0.1019049834 |
- |
- |
PREDICTED: protein WHI3 [Jatropha curcas] |
| 12 |
Hb_000292_100 |
0.1021292513 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 13 |
Hb_002303_020 |
0.1021738732 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 14 |
Hb_003929_070 |
0.1022030875 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 15 |
Hb_171900_090 |
0.1022988601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_062226_130 |
0.1028467144 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 17 |
Hb_000731_200 |
0.1035570365 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002157_120 |
0.1050647076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008225_090 |
0.1050828138 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000130_270 |
0.1052378468 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |