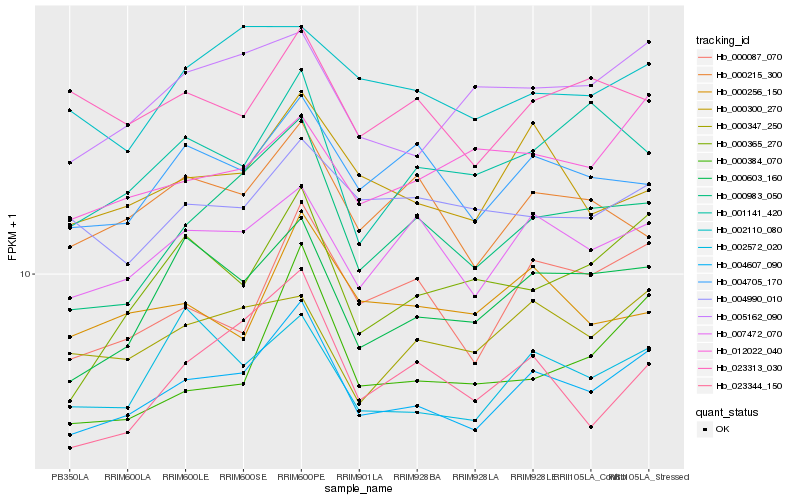
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004607_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 2 |
Hb_000983_050 |
0.0701133814 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_007472_070 |
0.0710474807 |
- |
- |
cir, putative [Ricinus communis] |
| 4 |
Hb_000603_160 |
0.0749872938 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 5 |
Hb_000087_070 |
0.077408192 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 6 |
Hb_000300_270 |
0.0793056874 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_005162_090 |
0.0799936394 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |
| 8 |
Hb_004705_170 |
0.080321274 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 9 |
Hb_002572_020 |
0.0833958628 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 10 |
Hb_000347_250 |
0.0868576917 |
- |
- |
PREDICTED: uncharacterized protein LOC105133971 [Populus euphratica] |
| 11 |
Hb_000365_270 |
0.0901689815 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 12 |
Hb_002110_080 |
0.0902388291 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 13 |
Hb_000384_070 |
0.0904283942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_023344_150 |
0.0909122171 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 15 |
Hb_001141_420 |
0.0909947677 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 16 |
Hb_023313_030 |
0.0917498801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004990_010 |
0.0917666459 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 18 |
Hb_012022_040 |
0.0919383742 |
- |
- |
Protein SIS1, putative [Ricinus communis] |
| 19 |
Hb_000215_300 |
0.092114292 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_000256_150 |
0.0928061516 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |