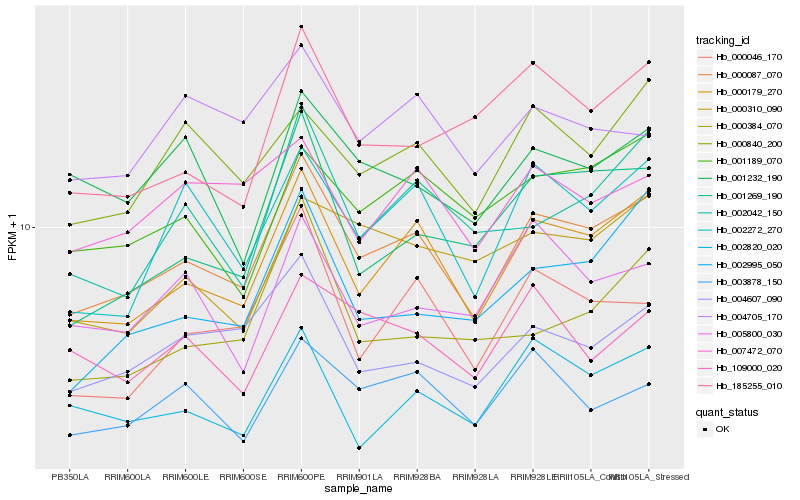
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_070 |
0.0 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 2 |
Hb_000179_270 |
0.0431035835 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |
| 3 |
Hb_002272_270 |
0.0763260433 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 4 |
Hb_004607_090 |
0.077408192 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 5 |
Hb_001269_190 |
0.079798755 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 6 |
Hb_000840_200 |
0.0799089888 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_003878_150 |
0.0802462482 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_000046_170 |
0.0809057991 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001189_070 |
0.083176443 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 10 |
Hb_002820_020 |
0.0837306925 |
- |
- |
hypothetical protein VITISV_005587 [Vitis vinifera] |
| 11 |
Hb_002042_150 |
0.0860051937 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 12 |
Hb_007472_070 |
0.0877426385 |
- |
- |
cir, putative [Ricinus communis] |
| 13 |
Hb_002995_050 |
0.0885549924 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000310_090 |
0.0899154008 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_185255_010 |
0.0899640156 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 16 |
Hb_109000_020 |
0.090446431 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 17 |
Hb_005800_030 |
0.0927150416 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 18 |
Hb_001232_190 |
0.093030673 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000384_070 |
0.0933961169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004705_170 |
0.0934154411 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |