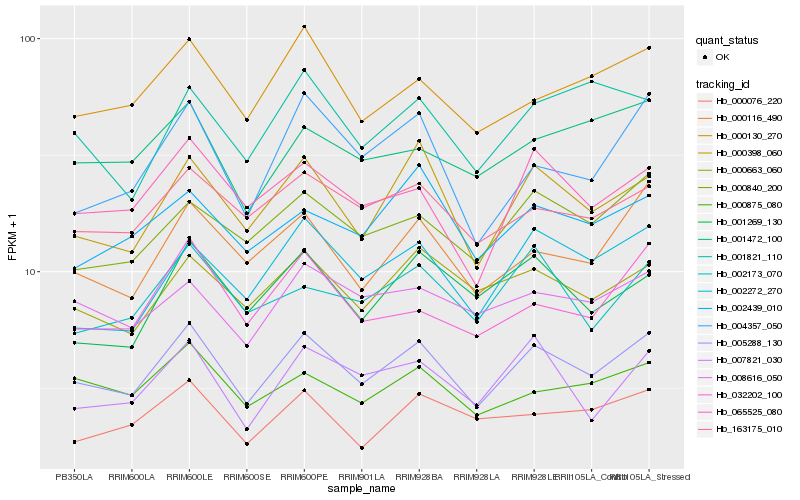
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_130 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_000398_060 |
0.0675941936 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 3 |
Hb_000116_490 |
0.0780492358 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 4 |
Hb_163175_010 |
0.0789162093 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 5 |
Hb_000840_200 |
0.0800984581 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_065525_080 |
0.0803248716 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 7 |
Hb_000663_060 |
0.0805532101 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 8 |
Hb_002272_270 |
0.0811428655 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 9 |
Hb_000130_270 |
0.0835688146 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000875_080 |
0.0846276247 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 11 |
Hb_007821_030 |
0.0874591238 |
- |
- |
5'-nucleotidase domain-containing [Gossypium arboreum] |
| 12 |
Hb_008616_050 |
0.0877063915 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 13 |
Hb_002173_070 |
0.0883993821 |
- |
- |
MRG family protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_002439_010 |
0.0889714835 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 15 |
Hb_001269_130 |
0.0894665418 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 16 |
Hb_004357_050 |
0.0896616735 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 17 |
Hb_032202_100 |
0.0901891929 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 18 |
Hb_000076_220 |
0.0902318373 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001472_100 |
0.0925864151 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 20 |
Hb_001821_110 |
0.0951979367 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |