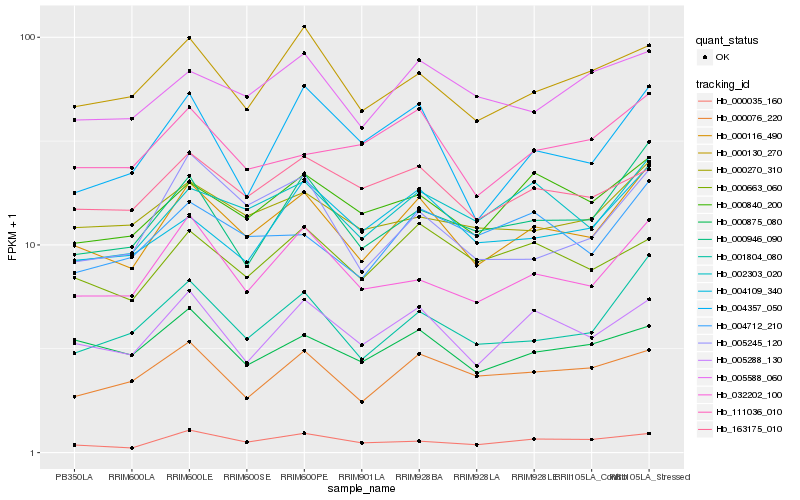
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_490 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 2 |
Hb_001804_080 |
0.0730034998 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 3 |
Hb_005288_130 |
0.0780492358 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_032202_100 |
0.0798692767 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 5 |
Hb_000946_090 |
0.0814443541 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_002303_020 |
0.0862028501 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 7 |
Hb_000270_310 |
0.088959313 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 8 |
Hb_000130_270 |
0.0899804847 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000840_200 |
0.0918936916 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_004109_340 |
0.0934085052 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14-like [Jatropha curcas] |
| 11 |
Hb_000035_160 |
0.0943276881 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 12 |
Hb_004712_210 |
0.0954878726 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Jatropha curcas] |
| 13 |
Hb_005588_060 |
0.0961768461 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 14 |
Hb_111036_010 |
0.0969292349 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 15 |
Hb_163175_010 |
0.0971126556 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 16 |
Hb_005245_120 |
0.0972857427 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_004357_050 |
0.0991519149 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 18 |
Hb_000663_060 |
0.0992275477 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 19 |
Hb_000076_220 |
0.0993697933 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000875_080 |
0.0994956073 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |