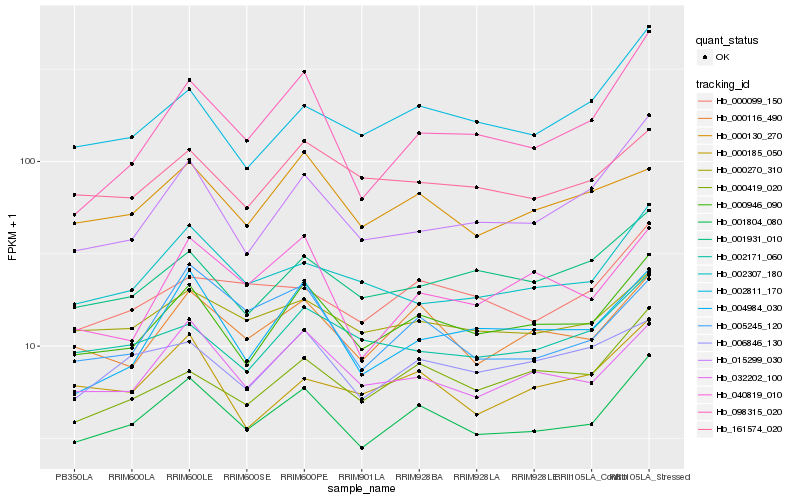
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001804_080 |
0.0 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 2 |
Hb_000946_090 |
0.0615837688 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000116_490 |
0.0730034998 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 4 |
Hb_032202_100 |
0.0819418371 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 5 |
Hb_000270_310 |
0.0907919955 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 6 |
Hb_002307_180 |
0.091871487 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_005245_120 |
0.0923444027 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000099_150 |
0.0969587354 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 9 |
Hb_000419_020 |
0.0986769146 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001931_010 |
0.0989863023 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 11 |
Hb_006846_130 |
0.0998095295 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 1-like [Jatropha curcas] |
| 12 |
Hb_161574_020 |
0.10033137 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 13 |
Hb_004984_030 |
0.1005453928 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_015299_030 |
0.101292325 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 15 |
Hb_002811_170 |
0.1015049799 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 16 |
Hb_000185_050 |
0.1015478628 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 17 |
Hb_000130_270 |
0.1030452115 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002171_060 |
0.1045329297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_098315_020 |
0.1056975893 |
- |
- |
hypothetical protein M569_15712 [Genlisea aurea] |
| 20 |
Hb_040819_010 |
0.1067522915 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |