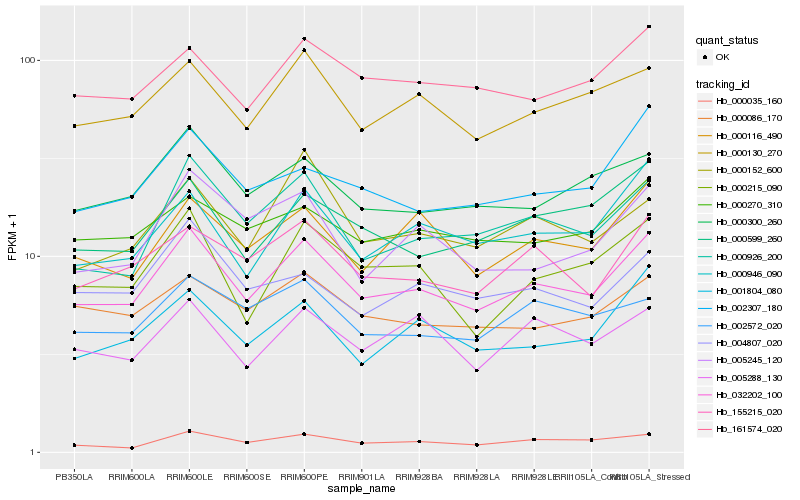
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032202_100 |
0.0 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 2 |
Hb_000130_270 |
0.0722914061 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000926_200 |
0.0743432201 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_155215_020 |
0.075398351 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Populus euphratica] |
| 5 |
Hb_000035_160 |
0.0778797081 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 6 |
Hb_000946_090 |
0.0787726902 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_161574_020 |
0.0798102575 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 8 |
Hb_000116_490 |
0.0798692767 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 9 |
Hb_000300_260 |
0.0798856429 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 10 |
Hb_001804_080 |
0.0819418371 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 11 |
Hb_000215_090 |
0.084285931 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000086_170 |
0.0847920568 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 13 |
Hb_000599_260 |
0.0848755744 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 14 |
Hb_000270_310 |
0.0862691172 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 15 |
Hb_002307_180 |
0.0888215353 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005288_130 |
0.0901891929 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_002572_020 |
0.0904612662 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 18 |
Hb_005245_120 |
0.0909623828 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000152_600 |
0.0914827067 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 20 |
Hb_004807_020 |
0.0919705117 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |