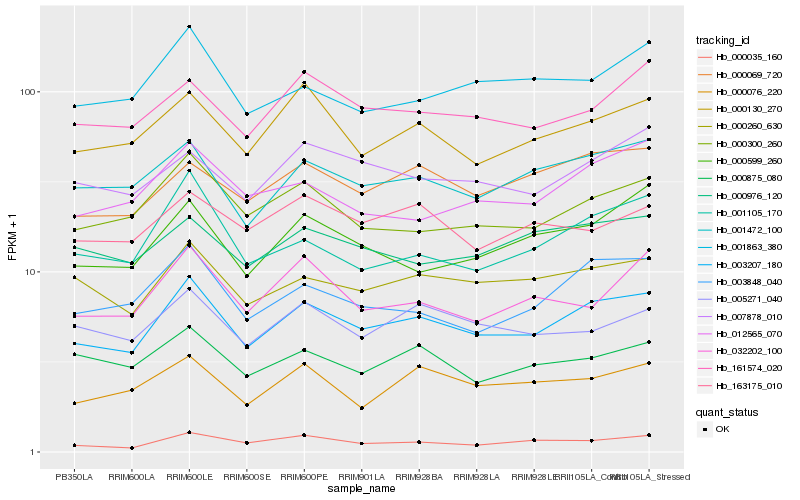
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003207_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001472_100 |
0.0736331379 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 3 |
Hb_000260_630 |
0.0757318524 |
- |
- |
PREDICTED: altered inheritance rate of mitochondria protein 25 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000300_260 |
0.0794145001 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 5 |
Hb_003848_040 |
0.0796778655 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_000069_720 |
0.0823528204 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 7 |
Hb_000130_270 |
0.0829293711 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000035_160 |
0.0865965585 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 9 |
Hb_161574_020 |
0.0874499604 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_007878_010 |
0.0886400472 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 11 |
Hb_001105_170 |
0.0889048405 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000599_260 |
0.0909944981 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 13 |
Hb_005271_040 |
0.0916319957 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 14 |
Hb_000076_220 |
0.0920527396 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_012565_070 |
0.0920802077 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 16 |
Hb_000875_080 |
0.0925074593 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 17 |
Hb_001863_380 |
0.0931710374 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 18 |
Hb_000976_120 |
0.0933889432 |
- |
- |
PREDICTED: DNA polymerase zeta processivity subunit [Jatropha curcas] |
| 19 |
Hb_032202_100 |
0.0936511144 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 20 |
Hb_163175_010 |
0.0980841572 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |