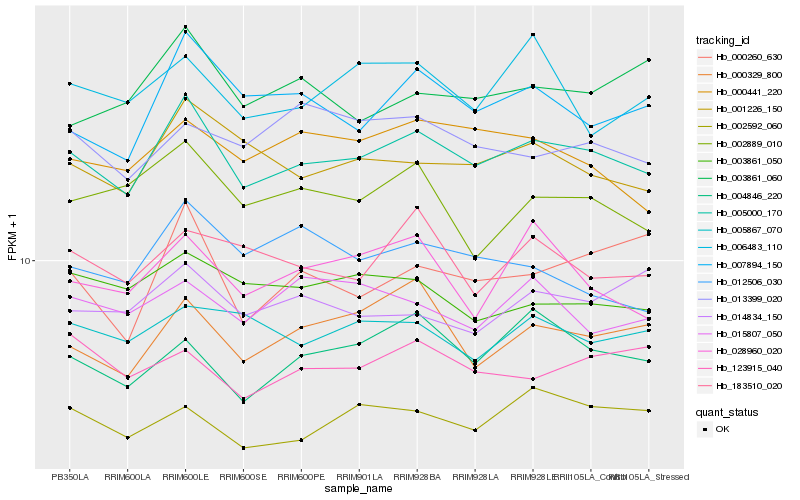
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_170 |
0.0 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 2 |
Hb_001226_150 |
0.0569984583 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_007894_150 |
0.0605085095 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 4 |
Hb_000329_800 |
0.0652715186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004846_220 |
0.0667328355 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002592_060 |
0.0674645267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000441_220 |
0.0684629315 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002889_010 |
0.06981731 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 9 |
Hb_183510_020 |
0.0704105302 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 10 |
Hb_003861_050 |
0.0704533119 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 11 |
Hb_005867_070 |
0.070916655 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_028960_020 |
0.071327903 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003861_060 |
0.0736585946 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 14 |
Hb_123915_040 |
0.073722024 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 15 |
Hb_000260_630 |
0.0738051387 |
- |
- |
PREDICTED: altered inheritance rate of mitochondria protein 25 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006483_110 |
0.0739684094 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 17 |
Hb_013399_020 |
0.0751017878 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 18 |
Hb_015807_050 |
0.0755476345 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 19 |
Hb_012506_030 |
0.075586824 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 20 |
Hb_014834_150 |
0.0759068864 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |