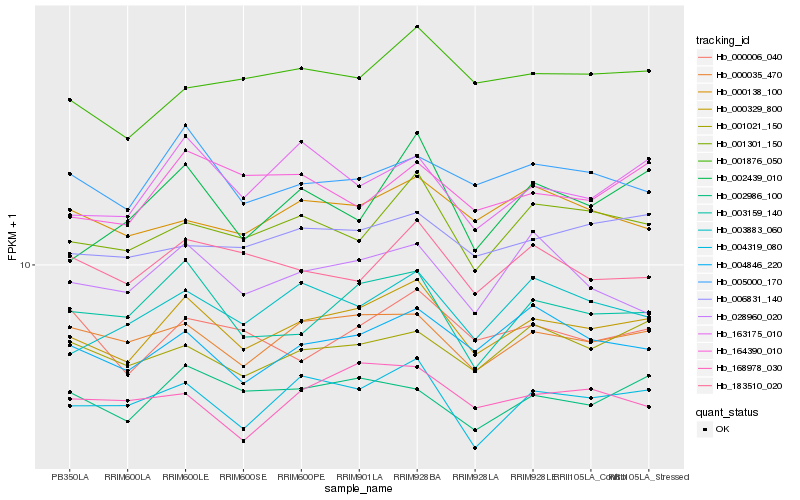
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_800 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004319_080 |
0.0578361447 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 3 |
Hb_000035_470 |
0.0595542794 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001301_150 |
0.0624628827 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 5 |
Hb_004846_220 |
0.0630919341 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003159_140 |
0.065059163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005000_170 |
0.0652715186 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 8 |
Hb_001876_050 |
0.0672707093 |
- |
- |
hypothetical protein JCGZ_01286 [Jatropha curcas] |
| 9 |
Hb_003883_060 |
0.0690414758 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_163175_010 |
0.0697814393 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 11 |
Hb_002986_100 |
0.0698344373 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 12 |
Hb_006831_140 |
0.0704132341 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000138_100 |
0.0707154851 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 14 |
Hb_001021_150 |
0.0713770218 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 15 |
Hb_183510_020 |
0.0728484767 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 16 |
Hb_168978_030 |
0.0745976652 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 17 |
Hb_000006_040 |
0.0746246324 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_164390_010 |
0.0746980442 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 19 |
Hb_028960_020 |
0.0754671197 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002439_010 |
0.0773249859 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |