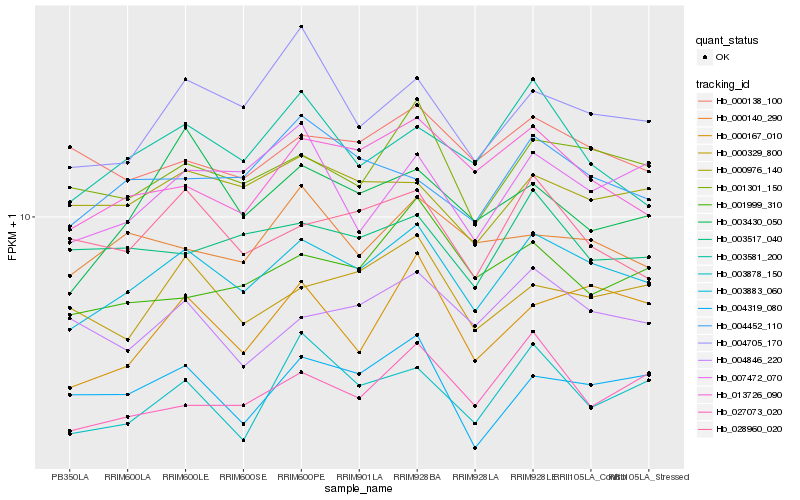
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003883_060 |
0.0 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001301_150 |
0.0551033921 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 3 |
Hb_013726_090 |
0.0613788095 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 4 |
Hb_004705_170 |
0.0674354009 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 5 |
Hb_000976_140 |
0.0675584196 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003581_200 |
0.0682033315 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 7 |
Hb_000167_010 |
0.068488382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004452_110 |
0.0687066656 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004319_080 |
0.0688398912 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 10 |
Hb_000329_800 |
0.0690414758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_028960_020 |
0.0696533151 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000138_100 |
0.0700773472 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 13 |
Hb_027073_020 |
0.0701850633 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 14 |
Hb_000140_290 |
0.0708952199 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 15 |
Hb_003517_040 |
0.0717470661 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007472_070 |
0.0718418271 |
- |
- |
cir, putative [Ricinus communis] |
| 17 |
Hb_003878_150 |
0.071882722 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 18 |
Hb_004846_220 |
0.0723118049 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003430_050 |
0.0723279551 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 20 |
Hb_001999_310 |
0.0731596077 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |