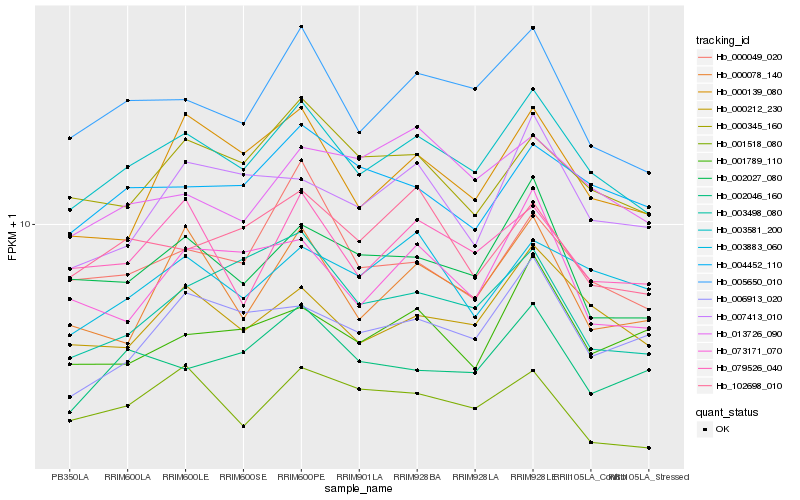
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 2 |
Hb_005650_010 |
0.0483135535 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000212_230 |
0.0532836581 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 4 |
Hb_002027_080 |
0.0604575438 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 5 |
Hb_000345_160 |
0.063130839 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 6 |
Hb_079526_040 |
0.0638676626 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 7 |
Hb_001518_080 |
0.0655260886 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 8 |
Hb_004452_110 |
0.0659032351 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000139_080 |
0.067990241 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003883_060 |
0.0682033315 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002046_160 |
0.068513656 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_102698_010 |
0.069418126 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_073171_070 |
0.0701295623 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 14 |
Hb_006913_020 |
0.070222654 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000049_020 |
0.070259373 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000078_140 |
0.0714472339 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 17 |
Hb_013726_090 |
0.0730744944 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 18 |
Hb_003498_080 |
0.0730917263 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_007413_010 |
0.0734046221 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 20 |
Hb_001789_110 |
0.0744962097 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |