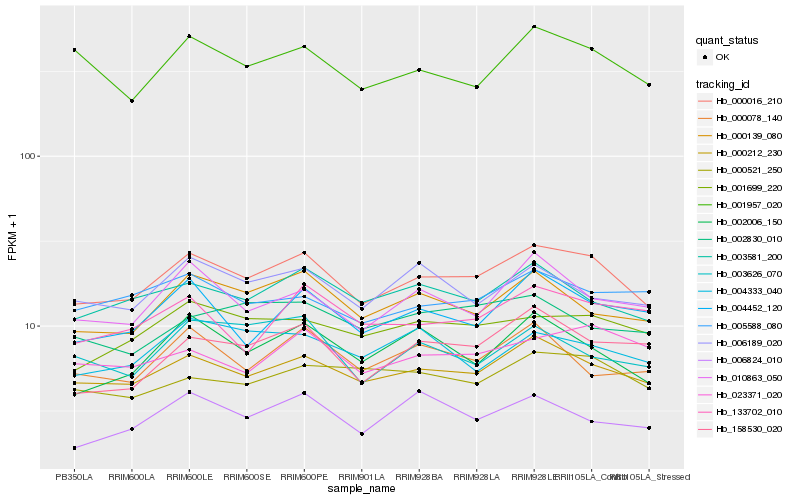
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_210 |
0.0 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 2 |
Hb_000212_230 |
0.0552092905 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 3 |
Hb_003581_200 |
0.0768639385 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 4 |
Hb_000139_080 |
0.0777082517 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001699_220 |
0.0832360814 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 6 |
Hb_002006_150 |
0.0862220125 |
- |
- |
copine, putative [Ricinus communis] |
| 7 |
Hb_001957_020 |
0.0862921329 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 8 |
Hb_010863_050 |
0.0879098445 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 9 |
Hb_004452_120 |
0.0888481675 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 10 |
Hb_006189_020 |
0.0914209954 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 11 |
Hb_000521_250 |
0.0915700012 |
- |
- |
PREDICTED: protein N-terminal asparagine amidohydrolase isoform X2 [Jatropha curcas] |
| 12 |
Hb_158530_020 |
0.0917937437 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 13 |
Hb_023371_020 |
0.0935278551 |
- |
- |
PREDICTED: uncharacterized protein LOC105633512 [Jatropha curcas] |
| 14 |
Hb_000078_140 |
0.0945151746 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 15 |
Hb_133702_010 |
0.0950897523 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_004333_040 |
0.0962138085 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 17 |
Hb_005588_080 |
0.0964551937 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 18 |
Hb_002830_010 |
0.0973740189 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_006824_010 |
0.0976820651 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_003626_070 |
0.0978305236 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |