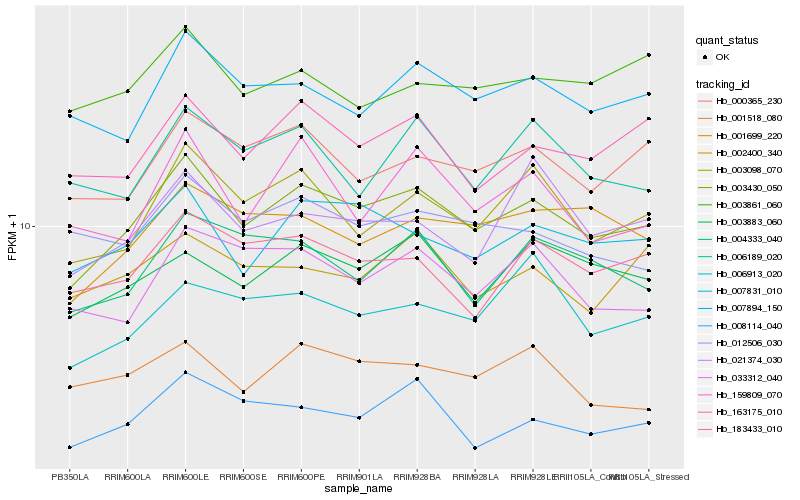
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003430_050 |
0.0 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 2 |
Hb_007831_010 |
0.0643014342 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 3 |
Hb_003098_070 |
0.0648304075 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 4 |
Hb_000365_230 |
0.0720249417 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 5 |
Hb_163175_010 |
0.0721403626 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 6 |
Hb_003883_060 |
0.0723279551 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007894_150 |
0.0726334089 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 8 |
Hb_001699_220 |
0.0728599013 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 9 |
Hb_183433_010 |
0.0758967013 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 10 |
Hb_008114_040 |
0.0760541169 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 11 |
Hb_004333_040 |
0.0773573384 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 12 |
Hb_033312_040 |
0.0790640384 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001518_080 |
0.0823195166 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 14 |
Hb_021374_030 |
0.0827156007 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 15 |
Hb_159809_070 |
0.0828128136 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 16 |
Hb_006189_020 |
0.0831370668 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 17 |
Hb_002400_340 |
0.0832888292 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 18 |
Hb_012506_030 |
0.0833570294 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 19 |
Hb_003861_060 |
0.0834418497 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 20 |
Hb_006913_020 |
0.0835695386 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |