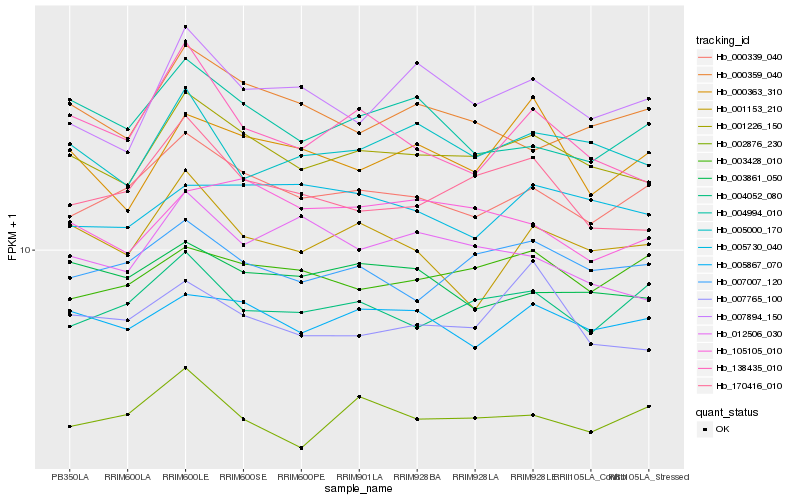
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001226_150 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_170416_010 |
0.0544745068 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 3 |
Hb_005867_070 |
0.0564489344 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_005000_170 |
0.0569984583 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 5 |
Hb_000339_040 |
0.0579252749 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_007007_120 |
0.0591091138 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007894_150 |
0.0628073881 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 8 |
Hb_138435_010 |
0.0642198898 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 9 |
Hb_003861_050 |
0.0642452982 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 10 |
Hb_105105_010 |
0.0645204186 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 11 |
Hb_007765_100 |
0.0656030308 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 12 |
Hb_012506_030 |
0.0659364778 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 13 |
Hb_002876_230 |
0.0660973933 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 14 |
Hb_000359_040 |
0.0672173912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000363_310 |
0.0686859394 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 16 |
Hb_004994_010 |
0.0687850549 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 17 |
Hb_005730_040 |
0.0689946272 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 18 |
Hb_003428_010 |
0.0693013363 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 19 |
Hb_004052_080 |
0.0695217736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001153_210 |
0.0698655946 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |