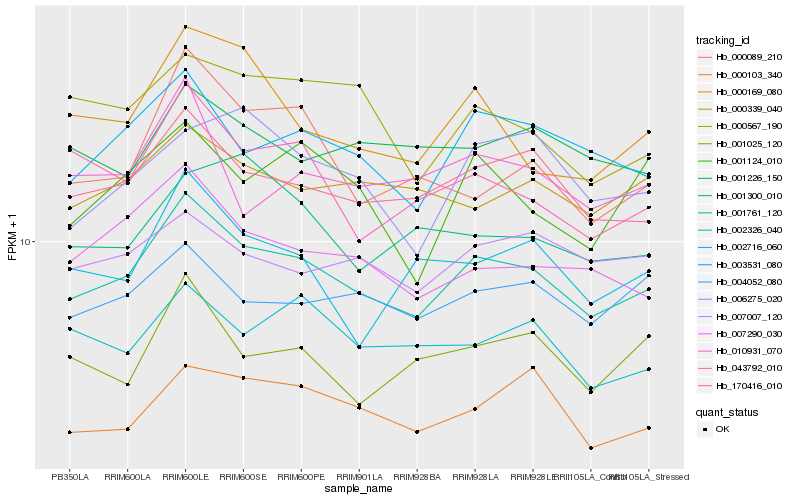
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001761_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 2 |
Hb_170416_010 |
0.0534643279 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 3 |
Hb_003531_080 |
0.0628657986 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007290_030 |
0.0700913961 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 5 |
Hb_004052_080 |
0.0733166536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000339_040 |
0.0736560592 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_000103_340 |
0.0738166927 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 8 |
Hb_043792_010 |
0.0757634551 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 9 |
Hb_007007_120 |
0.077698976 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000169_080 |
0.0778259643 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 11 |
Hb_002326_040 |
0.0786342341 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 12 |
Hb_002716_060 |
0.0797805044 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 13 |
Hb_001226_150 |
0.0803706574 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_001025_120 |
0.0804879325 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_000567_190 |
0.0805623053 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_010931_070 |
0.0807363227 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000089_210 |
0.0827001794 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_006275_020 |
0.0832421403 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 19 |
Hb_001124_010 |
0.0835986924 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001300_010 |
0.0848851551 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |