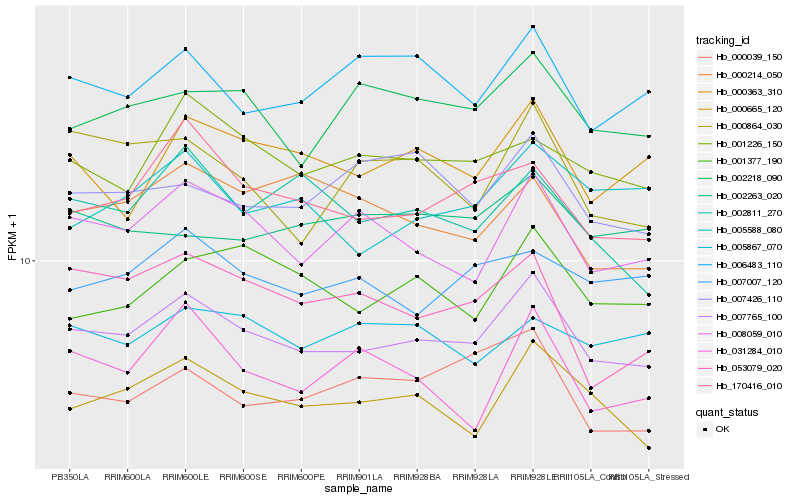
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_100 |
0.0 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 2 |
Hb_000665_120 |
0.0543279833 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 3 |
Hb_008059_010 |
0.0565267476 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 4 |
Hb_170416_010 |
0.0602739709 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 5 |
Hb_002218_090 |
0.0649406123 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 6 |
Hb_000363_310 |
0.0651042484 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 7 |
Hb_001226_150 |
0.0656030308 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_006483_110 |
0.0671426737 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 9 |
Hb_001377_190 |
0.0685615697 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007007_120 |
0.0687461444 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000039_150 |
0.0700219535 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 12 |
Hb_053079_020 |
0.0712152908 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 13 |
Hb_002811_270 |
0.0713199617 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 14 |
Hb_005588_080 |
0.0717948362 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 15 |
Hb_000864_030 |
0.0728265793 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007426_110 |
0.0733513152 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 17 |
Hb_005867_070 |
0.0739120362 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_031284_010 |
0.0740849879 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 19 |
Hb_002263_020 |
0.0745275715 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 20 |
Hb_000214_050 |
0.0768022998 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |