| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
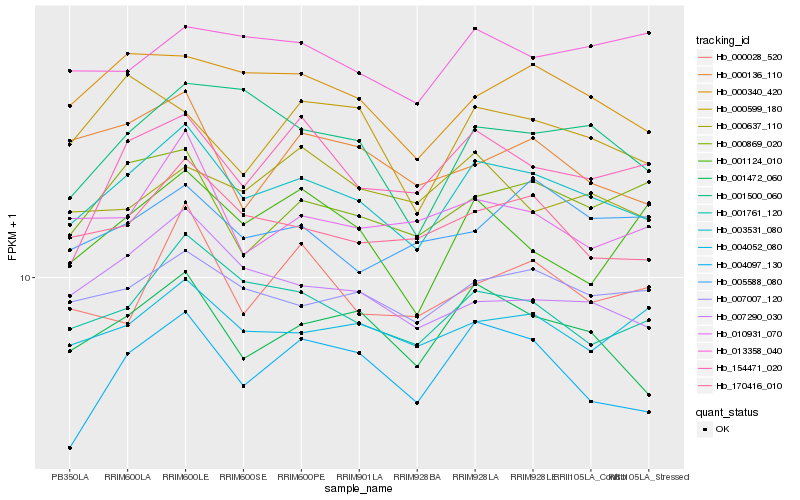
| 1 |
Hb_003531_080 |
0.0 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001761_120 |
0.0628657986 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 3 |
Hb_001472_060 |
0.0638531664 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_170416_010 |
0.0650445954 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 5 |
Hb_007007_120 |
0.066583076 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000340_420 |
0.0702627265 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_007290_030 |
0.0780069422 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 8 |
Hb_004052_080 |
0.0781271073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000869_020 |
0.0809109119 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 10 |
Hb_001500_060 |
0.080995148 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |
| 11 |
Hb_010931_070 |
0.0819036035 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004097_130 |
0.0855744149 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 13 |
Hb_154471_020 |
0.0859895874 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 14 |
Hb_013358_040 |
0.0886823744 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 15 |
Hb_000136_110 |
0.0888301836 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 16 |
Hb_000028_520 |
0.0915844738 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 17 |
Hb_005588_080 |
0.091746334 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 18 |
Hb_000599_180 |
0.0939685884 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_001124_010 |
0.0941211832 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000637_110 |
0.0941412879 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |