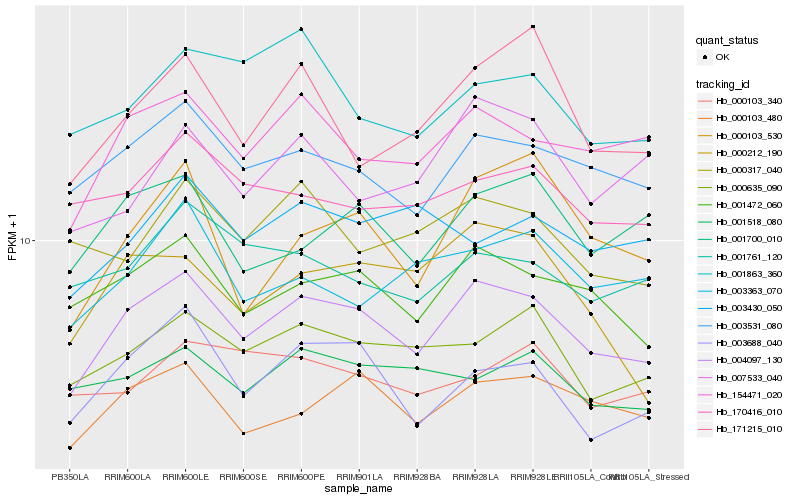
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004097_130 |
0.0 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 2 |
Hb_001472_060 |
0.084045369 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003531_080 |
0.0855744149 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_154471_020 |
0.0876068378 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 5 |
Hb_000103_480 |
0.09291542 |
- |
- |
- |
| 6 |
Hb_000635_090 |
0.0962550261 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 7 |
Hb_003688_040 |
0.1059491072 |
- |
- |
- |
| 8 |
Hb_001700_010 |
0.1092018581 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 9 |
Hb_171215_010 |
0.1123046091 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 10 |
Hb_170416_010 |
0.1133346548 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 11 |
Hb_000212_190 |
0.1133560329 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001518_080 |
0.1135301125 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 13 |
Hb_001761_120 |
0.1136239636 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 14 |
Hb_000317_040 |
0.1144922669 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_000103_530 |
0.1150795029 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 16 |
Hb_003363_070 |
0.1164286535 |
- |
- |
- |
| 17 |
Hb_001863_360 |
0.1173688305 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 18 |
Hb_007533_040 |
0.1181363168 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_000103_340 |
0.120315331 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 20 |
Hb_003430_050 |
0.1204933477 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |