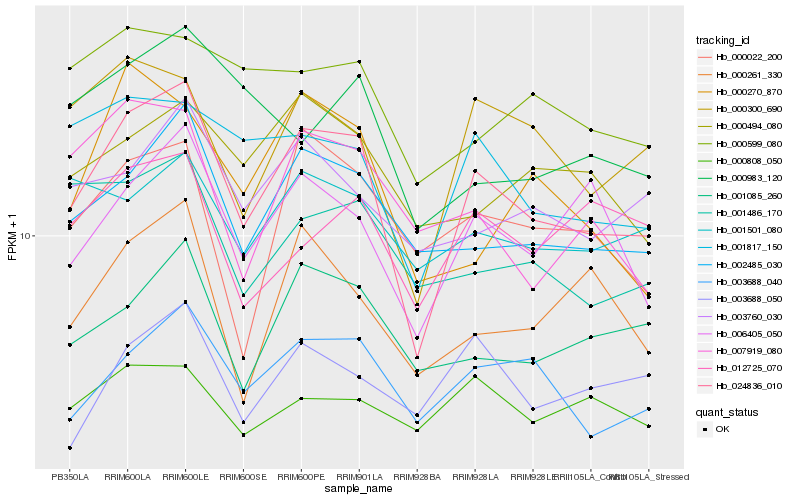
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024836_010 |
0.0 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 2 |
Hb_002485_030 |
0.1238164759 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 3 |
Hb_003688_040 |
0.1256714249 |
- |
- |
- |
| 4 |
Hb_000300_690 |
0.1323632685 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 5 |
Hb_006405_050 |
0.1326121988 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 6 |
Hb_001817_150 |
0.1358381764 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_001085_260 |
0.1423551833 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 8 |
Hb_003760_030 |
0.1518457648 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 9 |
Hb_001486_170 |
0.1520726435 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 10 |
Hb_000022_200 |
0.1523119517 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 11 |
Hb_000261_330 |
0.1526299822 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 12 |
Hb_007919_080 |
0.1544348009 |
- |
- |
PREDICTED: uncharacterized protein At1g76660 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000494_080 |
0.1548465511 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000270_870 |
0.1565226792 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 15 |
Hb_000599_080 |
0.1567995275 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 16 |
Hb_001501_080 |
0.1576288874 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012725_070 |
0.159237963 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000983_120 |
0.1596187646 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 19 |
Hb_003688_050 |
0.1596528221 |
- |
- |
PREDICTED: uncharacterized protein LOC105640616 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000808_050 |
0.1602494635 |
- |
- |
exonuclease, putative [Ricinus communis] |