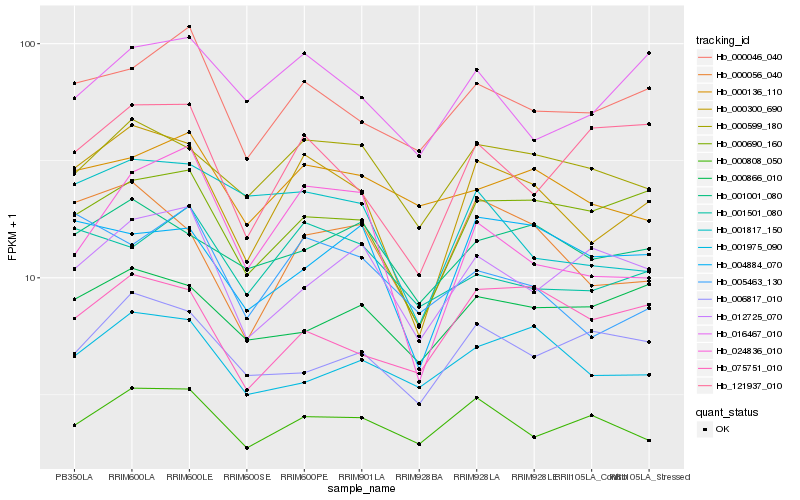
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_690 |
0.0 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 2 |
Hb_000056_040 |
0.1013643983 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 3 |
Hb_000690_160 |
0.1165077075 |
- |
- |
PREDICTED: uncharacterized protein LOC105121500 [Populus euphratica] |
| 4 |
Hb_000599_180 |
0.1232841527 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000046_040 |
0.1266637976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004884_070 |
0.1316899307 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 [Jatropha curcas] |
| 7 |
Hb_024836_010 |
0.1323632685 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 8 |
Hb_001817_150 |
0.1334620094 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_001001_080 |
0.135987122 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 10 |
Hb_075751_010 |
0.1390810846 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005463_130 |
0.1393966862 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 12 |
Hb_000808_050 |
0.1399816786 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 13 |
Hb_121937_010 |
0.1407126874 |
- |
- |
hypothetical protein B456_013G261900 [Gossypium raimondii] |
| 14 |
Hb_016467_010 |
0.1426520135 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 15 |
Hb_001501_080 |
0.1436104301 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001975_090 |
0.1453416488 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 17 |
Hb_000136_110 |
0.1474001667 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 18 |
Hb_006817_010 |
0.1475449195 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000866_010 |
0.1475638411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012725_070 |
0.1476130065 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |