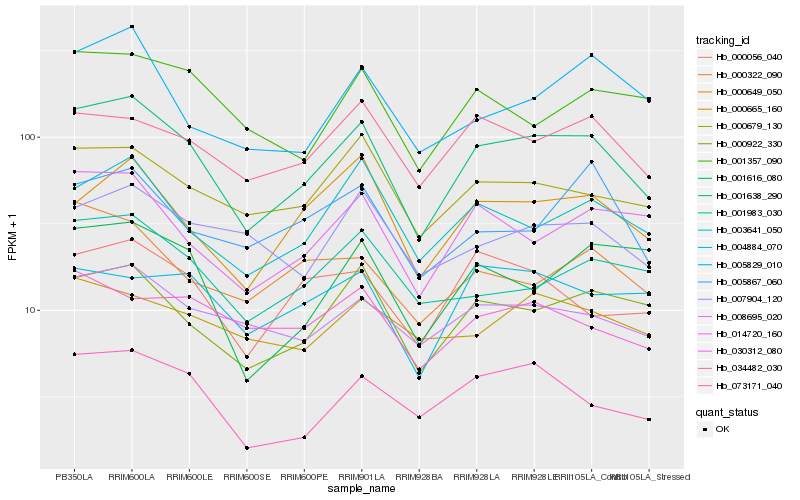
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_290 |
0.0 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000922_330 |
0.1027572657 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 3 |
Hb_003641_050 |
0.1114105088 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000679_130 |
0.1125080115 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_008695_020 |
0.1127502729 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 6 |
Hb_034482_030 |
0.1135253737 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_014720_160 |
0.1150871912 |
- |
- |
PREDICTED: anthranilate phosphoribosyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_073171_040 |
0.1167334574 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_000649_050 |
0.117680174 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001983_030 |
0.1210947209 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000056_040 |
0.1240934296 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 12 |
Hb_001357_090 |
0.127090957 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 13 |
Hb_005829_010 |
0.127779892 |
- |
- |
PREDICTED: uncharacterized protein LOC105640979 [Jatropha curcas] |
| 14 |
Hb_007904_120 |
0.1286652844 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_030312_080 |
0.1315917206 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 16 |
Hb_000322_090 |
0.1321108966 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_001616_080 |
0.1340333687 |
- |
- |
Oleoyl-acyl carrier protein thioesterase, putative [Ricinus communis] |
| 18 |
Hb_005867_060 |
0.1349352923 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase [Jatropha curcas] |
| 19 |
Hb_000665_160 |
0.1355591115 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 20 |
Hb_004884_070 |
0.1362592148 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 [Jatropha curcas] |